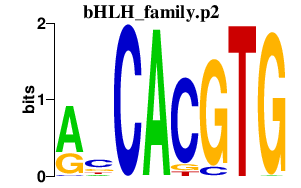
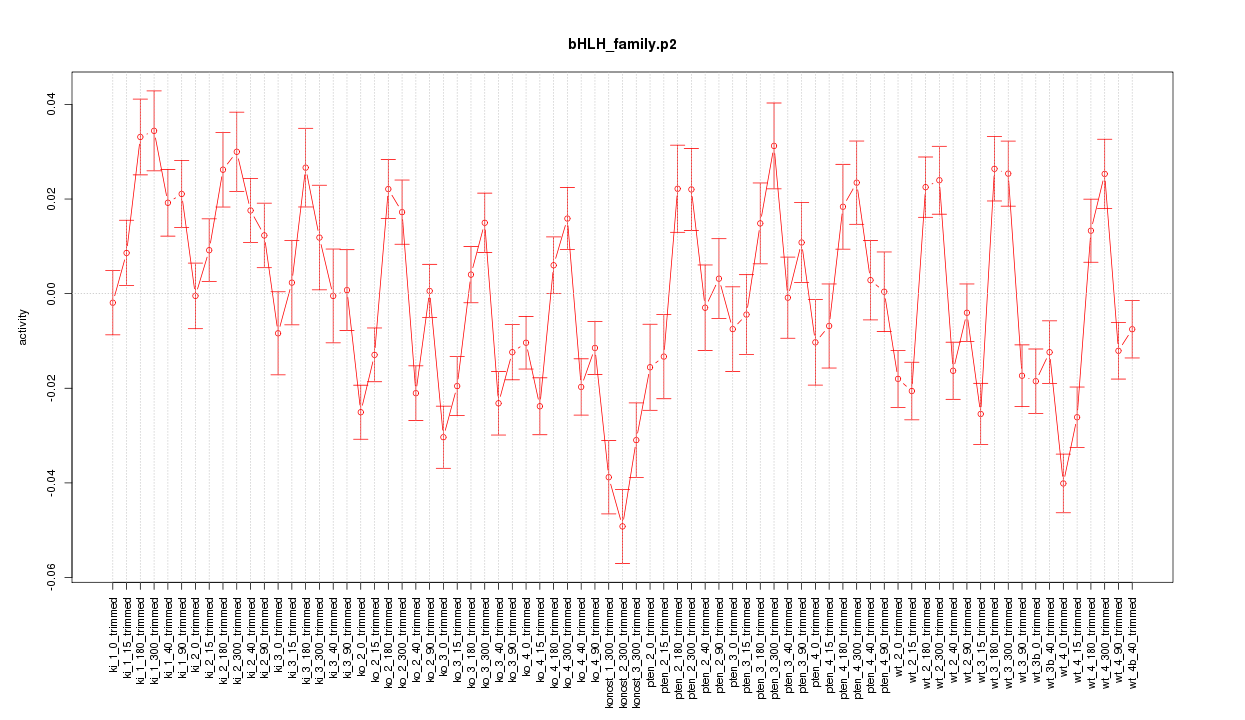
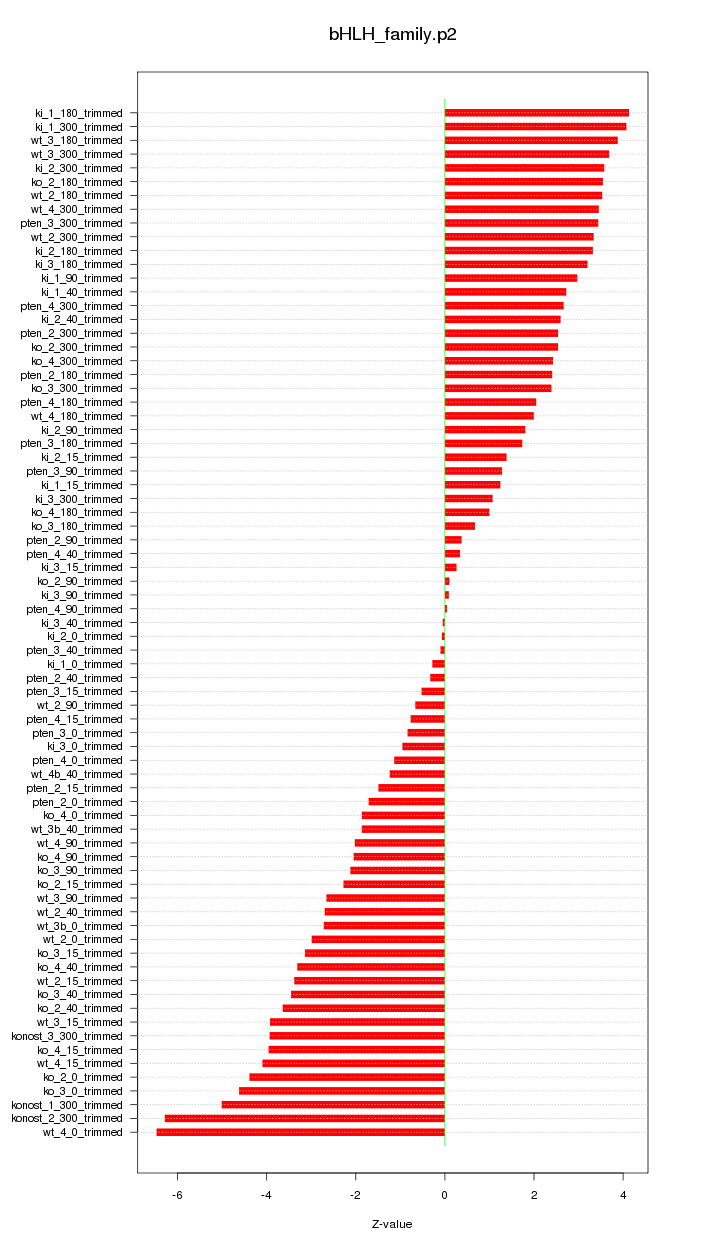
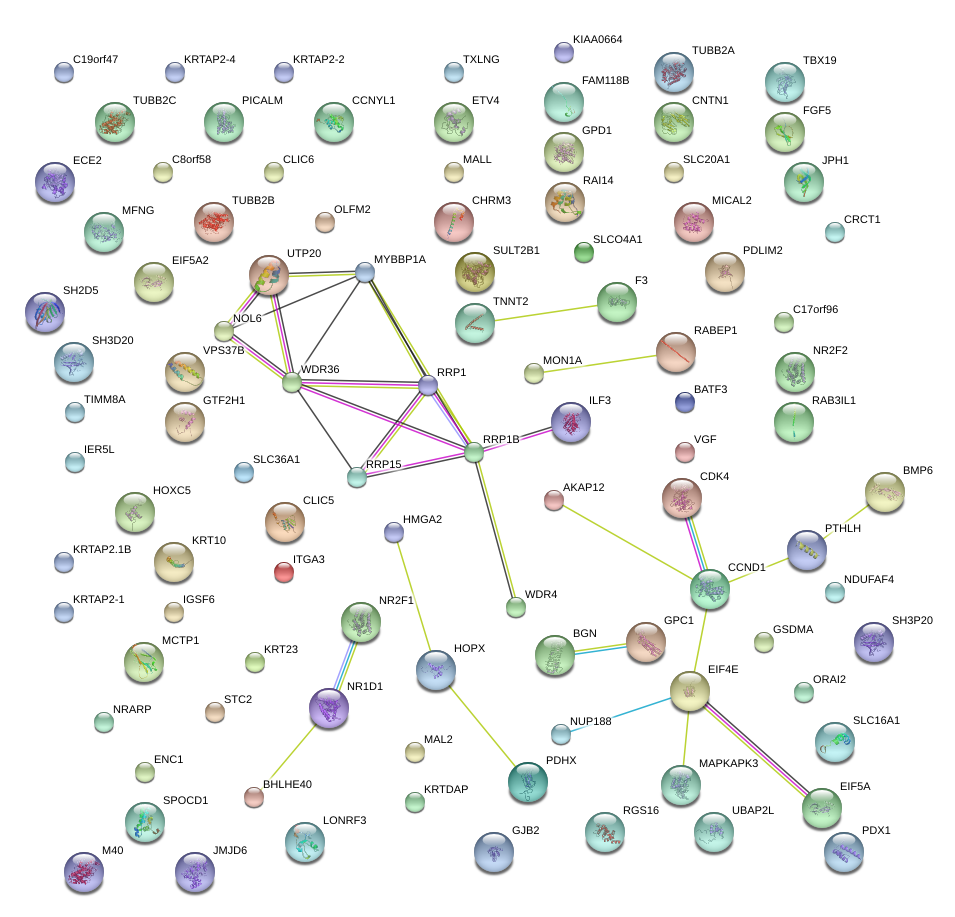
|
chr1_-_212873104
|
22.474
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr3_-_170626425
|
21.730
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_-_170626381
|
19.224
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr11_-_61684981
|
19.125
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr2_+_113403433
|
18.569
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr13_-_20767036
|
17.902
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr11_+_12132063
|
16.435
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_-_32281577
|
16.115
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr19_+_10764896
|
16.070
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr5_+_150827187
|
15.766
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr19_+_49055421
|
15.529
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr13_+_28494136
|
15.427
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr1_-_21058701
|
15.356
|
|
SH2D5
|
SH2 domain containing 5
|
|
chr12_-_58146110
|
15.182
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146108
|
14.969
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_38256548
|
14.710
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr5_-_172755195
|
14.676
|
|
STC2
|
stanniocalcin 2
|
|
chr1_-_212873306
|
14.604
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr12_-_58146025
|
14.408
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_-_85780105
|
13.878
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr5_+_150827138
|
13.663
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr6_+_151646634
|
13.637
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_-_212873197
|
13.477
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr19_+_10765106
|
13.459
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr9_-_131940481
|
13.331
|
|
IER5L
|
immediate early response 5-like
|
|
chr17_-_38256905
|
13.196
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr5_+_150827204
|
12.918
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr12_-_58146118
|
12.815
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_-_85780088
|
12.781
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_-_131940538
|
12.752
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr19_+_10764987
|
12.605
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr1_+_239549834
|
12.138
|
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr12_-_28122884
|
11.778
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr5_-_73937248
|
11.731
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_-_85779821
|
11.713
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr17_-_4458618
|
10.960
|
NM_001105538
NM_014520
|
MYBBP1A
|
MYB binding protein (P160) 1a
|
|
chr4_-_99850242
|
10.757
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr11_+_18344113
|
10.650
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr8_-_75233461
|
10.510
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr17_-_41623652
|
10.490
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chrX_+_118108575
|
10.468
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr2_+_241375207
|
10.331
|
|
GPC1
|
glypican 1
|
|
chr4_-_99850280
|
10.318
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chrX_+_16804531
|
10.289
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr19_-_10047014
|
10.280
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr6_+_7726268
|
10.178
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr6_-_45983279
|
10.065
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr9_-_33473846
|
10.053
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr1_+_154193294
|
10.012
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr9_-_33473876
|
10.009
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr11_-_2018568
|
9.948
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr5_+_34656482
|
9.845
|
|
RAI14
|
retinoic acid induced 14
|
|
chr21_-_44299605
|
9.800
|
|
WDR4
|
WD repeat domain 4
|
|
chr5_-_172755418
|
9.760
|
|
STC2
|
stanniocalcin 2
|
|
chr11_-_2018703
|
9.685
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_+_118108595
|
9.648
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr8_+_22436253
|
9.578
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr22_-_37882214
|
9.460
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr3_+_50654558
|
9.416
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr11_+_69455837
|
9.396
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr1_-_95007139
|
9.318
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr5_+_92920592
|
9.290
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chrX_+_152770083
|
9.262
|
|
BGN
|
biglycan
|
|
chr9_-_140196702
|
9.234
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr19_-_35981345
|
9.228
|
NM_001244847
NM_207392
|
KRTDAP
|
keratinocyte differentiation-associated protein
|
|
chr9_-_33473919
|
9.180
|
NM_022917
NM_139235
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr1_-_182573393
|
9.123
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr21_+_45209398
|
9.116
|
NM_003683
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr11_-_2018446
|
9.037
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr13_-_20767100
|
9.027
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr3_-_49967194
|
8.938
|
NM_001142501
NM_032355
|
MON1A
|
MON1 homolog A (yeast)
|
|
chr17_-_74722843
|
8.935
|
NM_001081461
NM_015167
|
JMJD6
|
jumonji domain containing 6
|
|
chr5_+_34656595
|
8.823
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr3_+_50654600
|
8.778
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr21_+_45209448
|
8.776
|
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr2_+_208576365
|
8.655
|
|
CCNYL1
|
cyclin Y-like 1
|
|
chr12_+_54426831
|
8.654
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr20_+_61273823
|
8.572
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr17_-_43510279
|
8.559
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chrX_-_100603956
|
8.546
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_-_113498615
|
8.524
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr12_+_50497790
|
8.512
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr3_+_50654589
|
8.493
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr6_-_97345747
|
8.440
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr17_+_5185551
|
8.363
|
NM_001083585
NM_004703
|
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1
|
|
chr12_+_101673895
|
8.356
|
NM_014503
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast)
|
|
chr3_+_5021141
|
8.324
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr8_+_120220554
|
8.309
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr3_+_5021096
|
8.292
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr19_-_40854283
|
8.247
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr5_+_110427869
|
8.219
|
NM_139281
|
WDR36
|
WD repeat domain 36
|
|
chr6_-_3227776
|
8.194
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chr2_-_75062214
|
8.186
|
|
|
|
|
chr2_-_110873533
|
8.183
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr3_+_183967414
|
8.167
|
NM_014693
NM_032331
|
ECE2
|
endothelin converting enzyme 2
|
|
chr2_-_110873353
|
8.160
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chrX_-_100603673
|
8.159
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr9_+_131709991
|
8.156
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr17_+_48133331
|
8.098
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr5_+_34656367
|
8.098
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr17_-_36831156
|
8.090
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr17_+_38119225
|
8.085
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr4_+_81187741
|
8.022
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr7_+_102074002
|
7.998
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr11_+_126081618
|
7.990
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr4_-_57522469
|
7.981
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr17_-_43510204
|
7.967
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr3_+_183967469
|
7.918
|
|
ECE2
|
endothelin converting enzyme 2
|
|
chr2_-_110873638
|
7.910
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr1_-_53164014
|
7.904
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr12_+_66218141
|
7.886
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr17_-_74722614
|
7.845
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr2_-_110874142
|
7.843
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr17_-_2614926
|
7.832
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr1_+_218458646
|
7.635
|
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae)
|
|
chr17_-_39211462
|
7.611
|
NM_033032
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr17_-_39093671
|
7.522
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr5_-_94620277
|
7.498
|
NM_024717
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr17_+_48133357
|
7.473
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr17_-_39203423
|
7.469
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr1_-_201346804
|
7.423
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr6_+_151561094
|
7.414
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_-_100808832
|
7.398
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr11_-_2018334
|
7.398
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr3_+_5021305
|
7.372
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr1_+_152486977
|
7.342
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr9_+_131709970
|
7.313
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr12_-_123380628
|
7.291
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr17_-_38978824
|
7.291
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr11_+_72929392
|
7.279
|
NM_176071
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr2_-_220118610
|
7.217
|
NM_006000
|
TUBA4A
|
tubulin, alpha 4a
|
|
chr20_+_2633177
|
7.198
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr1_-_21059132
|
7.198
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr6_-_47277354
|
7.135
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr10_-_120514277
|
7.116
|
|
|
|
|
chr5_+_176873795
|
7.075
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr19_-_40854410
|
7.068
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr7_+_48128939
|
7.042
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr17_-_78450247
|
7.010
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr1_+_26496382
|
7.008
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr2_+_88991166
|
6.991
|
NM_144563
|
RPIA
|
ribose 5-phosphate isomerase A
|
|
chr9_-_135545327
|
6.968
|
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31
|
|
chr15_+_32933869
|
6.943
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr1_-_201346783
|
6.932
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr4_-_4291889
|
6.859
|
NM_017816
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr16_-_103557
|
6.810
|
NM_016310
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa
|
|
chr14_-_71275732
|
6.788
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr9_-_34048887
|
6.772
|
NM_018449
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr4_-_4291817
|
6.765
|
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr11_+_18343794
|
6.763
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr3_-_50360109
|
6.746
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr11_+_72929606
|
6.746
|
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr4_+_75230856
|
6.725
|
NM_001432
|
EREG
|
epiregulin
|
|
chr15_-_75932509
|
6.665
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr15_+_89164522
|
6.664
|
NM_022767
|
AEN
|
apoptosis enhancing nuclease
|
|
chr6_+_41889234
|
6.660
|
|
|
|
|
chr6_+_44191276
|
6.629
|
NM_001078175
NM_001078176
NM_001078177
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr1_-_156675237
|
6.628
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr20_+_23331372
|
6.610
|
NM_013248
|
NXT1
|
NTF2-like export factor 1
|
|
chr9_-_34048869
|
6.609
|
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr16_+_56965747
|
6.590
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr2_-_208634051
|
6.543
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr19_-_2783332
|
6.533
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr17_-_73874056
|
6.477
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr4_-_4291731
|
6.455
|
NM_001145725
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr19_+_45909466
|
6.450
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr1_+_44440595
|
6.404
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_-_74722629
|
6.396
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr4_-_11430502
|
6.346
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr19_+_18530087
|
6.342
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr2_-_130956011
|
6.340
|
NM_207312
|
TUBA3E
|
tubulin, alpha 3e
|
|
chr7_-_2354050
|
6.339
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr11_+_72929302
|
6.331
|
NM_002564
NM_176072
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr1_+_44440623
|
6.322
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_-_39222126
|
6.317
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr4_-_57522059
|
6.310
|
|
HOPX
|
HOP homeobox
|
|
chr11_-_64645925
|
6.259
|
|
EHD1
|
EH-domain containing 1
|
|
chr11_+_67159421
|
6.253
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr8_+_67341244
|
6.251
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr19_+_45909868
|
6.235
|
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr20_+_39657461
|
6.211
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr11_+_45944155
|
6.196
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr11_+_120081669
|
6.168
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr7_-_105162551
|
6.163
|
NM_019042
|
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae)
|
|
chr7_-_124405029
|
6.160
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr11_-_65667860
|
6.152
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr8_+_22436641
|
6.145
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr13_-_50699775
|
6.131
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr21_-_44299660
|
6.114
|
NM_018669
NM_033661
|
WDR4
|
WD repeat domain 4
|
|
chr10_-_82049178
|
6.107
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr6_+_151561508
|
6.080
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_-_64646053
|
6.055
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr19_-_1237840
|
6.048
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr8_+_145582208
|
6.044
|
NM_001253816
NM_001253815
|
GPR172A
|
G protein-coupled receptor 172A
|
|
chr3_+_183967514
|
6.037
|
|
ECE2
|
endothelin converting enzyme 2
|
|
chr6_+_31783314
|
6.036
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr17_+_40719064
|
6.021
|
NM_170607
NM_198204
NM_198205
|
MLX
|
MAX-like protein X
|
|
chr13_+_37005966
|
5.985
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr6_+_31795395
|
5.945
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr6_-_47277640
|
5.926
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|