|
chr7_+_128470462
|
14.431
|
NM_001127487
NM_001458
|
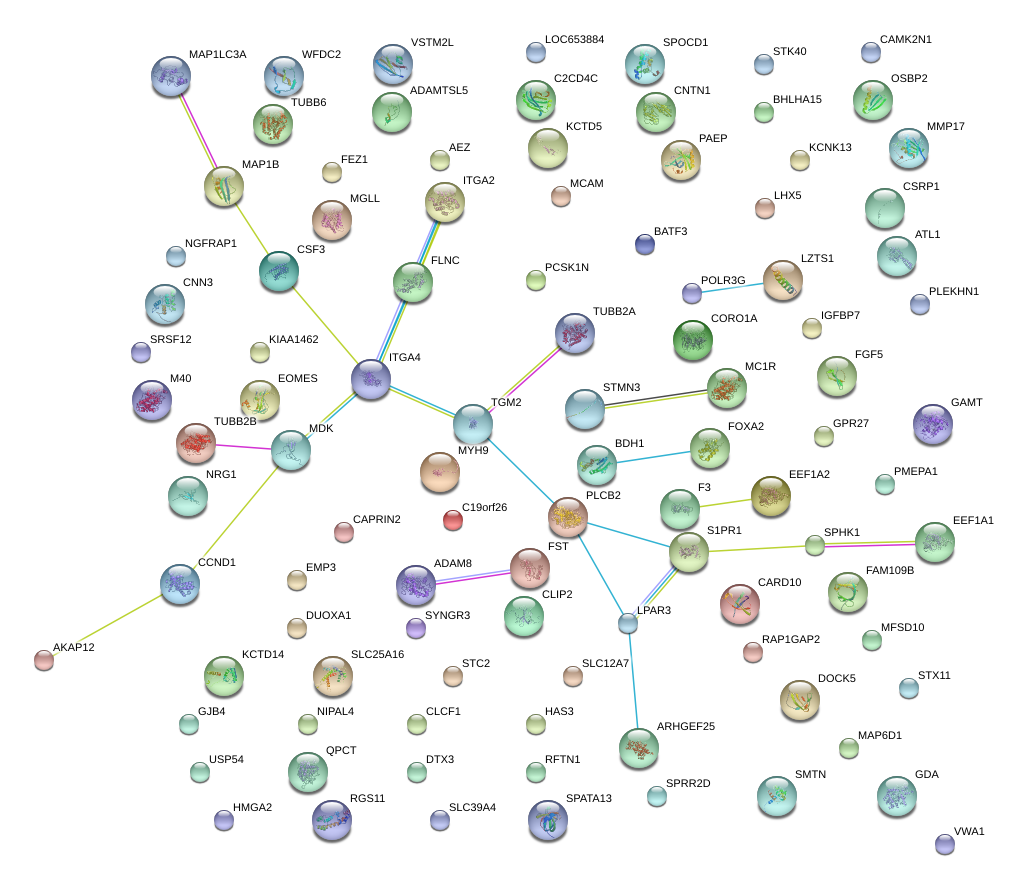
FLNC
|
filamin C, gamma
|
|
chr5_+_52776263
|
12.640
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr5_+_52776479
|
10.802
|
|
FST
|
follistatin
|
|
chr2_+_37571752
|
10.485
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr20_-_62284701
|
10.472
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr22_-_20137430
|
9.989
|
NM_001243537
|
LOC388849
|
uncharacterized LOC388849
|
|
chr1_+_35225341
|
8.402
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr4_+_81187741
|
8.310
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr5_+_52776408
|
7.827
|
|
FST
|
follistatin
|
|
chr16_+_30194925
|
7.149
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr10_-_30348433
|
7.071
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr20_+_33146500
|
6.915
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr19_-_1237840
|
6.699
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr16_+_30194730
|
6.590
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr4_-_57976516
|
6.574
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_57976550
|
6.461
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr6_+_144471631
|
6.188
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr5_+_156887204
|
6.132
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr12_+_58005217
|
5.921
|
NM_182947
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr3_-_127541160
|
5.911
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr20_+_44098392
|
5.910
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr17_+_74380676
|
5.715
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr1_-_20812271
|
5.613
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr11_-_119187811
|
5.600
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr5_+_156887026
|
5.333
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr1_+_1370902
|
5.183
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr12_+_66218141
|
5.167
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr22_+_31090792
|
5.152
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr15_-_45422056
|
5.141
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr1_-_95007139
|
5.105
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr19_+_48828820
|
5.014
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr12_-_30907447
|
4.995
|
NM_001002259
NM_001206856
NM_023925
NM_032156
|
CAPRIN2
|
caprin family member 2
|
|
chr9_+_74764266
|
4.966
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr16_+_69141442
|
4.834
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr6_+_151646634
|
4.745
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr8_+_32405727
|
4.716
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr5_+_71403040
|
4.706
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_27764197
|
4.685
|
|
EOMES
|
eomesodermin
|
|
chr4_-_57976355
|
4.554
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr11_+_46402333
|
4.553
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr19_-_1401474
|
4.444
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr20_-_22564893
|
4.440
|
|
FOXA2
|
forkhead box A2
|
|
chr12_+_132312917
|
4.403
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr12_+_57998600
|
4.357
|
NM_178502
|
DTX3
|
deltex homolog 3 (Drosophila)
|
|
chr20_-_62130390
|
4.166
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr17_+_2699731
|
4.148
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr22_-_37915209
|
4.030
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chrX_+_102632108
|
3.960
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr5_+_89770680
|
3.948
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr16_-_325867
|
3.925
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr8_+_25042066
|
3.911
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr19_-_409169
|
3.910
|
NM_001136263
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr5_-_1112106
|
3.907
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr3_+_71803200
|
3.900
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr7_+_73703704
|
3.870
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr3_-_183543295
|
3.869
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr13_+_24734788
|
3.849
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr17_+_38171613
|
3.825
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr14_+_90528107
|
3.793
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr1_-_95392501
|
3.771
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr1_+_901876
|
3.737
|
NM_001160184
NM_032129
|
PLEKHN1
|
pleckstrin homology domain containing, family N member 1
|
|
chr5_+_52284903
|
3.706
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr1_+_101702304
|
3.691
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr3_-_16555128
|
3.674
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr20_-_56285576
|
3.650
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr3_-_16554992
|
3.632
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr17_+_74380943
|
3.623
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr1_-_212873104
|
3.617
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr11_+_69455978
|
3.599
|
|
CCND1
|
cyclin D1
|
|
chr14_+_51026853
|
3.582
|
|
ATL1
|
atlastin GTPase 1
|
|
chr6_+_151561653
|
3.545
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr3_-_127541975
|
3.544
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr22_+_31477231
|
3.495
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr2_+_182321933
|
3.474
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr1_-_212873197
|
3.463
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr15_-_40600118
|
3.442
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_-_212873306
|
3.437
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr6_-_89827605
|
3.436
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr18_+_12308236
|
3.427
|
|
TUBB6
|
tubulin, beta 6 class V
|
|
chrX_-_48693934
|
3.420
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr1_-_153013593
|
3.416
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr11_-_77734285
|
3.379
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr11_-_67141647
|
3.378
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr20_+_36531548
|
3.362
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr4_-_775586
|
3.353
|
|
LOC100129917
|
uncharacterized LOC100129917
|
|
chr2_+_182322265
|
3.316
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr1_-_36851484
|
3.309
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr1_-_32281577
|
3.278
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr20_-_36793646
|
3.252
|
NM_004613
NM_198951
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr8_-_145638929
|
3.219
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr12_-_113909876
|
3.212
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr7_+_97841565
|
3.207
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr22_-_36783851
|
3.207
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr16_+_2039945
|
3.197
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr20_-_1165116
|
3.183
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr11_-_125366048
|
3.180
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr5_-_172756505
|
3.179
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr19_-_1513187
|
3.170
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr4_-_2935851
|
3.166
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr1_+_101702561
|
3.162
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_-_201475966
|
3.146
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr11_+_46403205
|
3.136
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr10_-_135090325
|
3.122
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr1_-_85358851
|
3.114
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr22_+_42470247
|
3.091
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr4_-_2936458
|
3.089
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr16_+_2732526
|
3.086
|
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr3_-_197282805
|
3.082
|
NM_004051
NM_203314
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr16_+_2732475
|
3.061
|
NM_018992
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr10_+_75532065
|
3.059
|
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr1_-_201476241
|
3.047
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr11_-_128392061
|
3.032
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr3_-_57199313
|
3.025
|
|
IL17RD
|
interleukin 17 receptor D
|
|
chr19_-_1512997
|
3.017
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr22_-_36783977
|
3.005
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr1_-_201476218
|
2.994
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr22_-_36783930
|
2.986
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr19_+_35521591
|
2.919
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr1_-_6546013
|
2.913
|
NM_001042665
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr3_-_185542807
|
2.893
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_+_16187266
|
2.885
|
|
TPM4
|
tropomyosin 4
|
|
chr20_+_35169876
|
2.881
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr22_-_45608346
|
2.872
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr19_+_48828627
|
2.862
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr9_+_33750463
|
2.857
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr19_+_16187306
|
2.836
|
|
TPM4
|
tropomyosin 4
|
|
chr17_-_62658311
|
2.820
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr8_-_145018057
|
2.806
|
NM_201382
|
PLEC
|
plectin
|
|
chr10_+_64133915
|
2.806
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr16_+_82660573
|
2.786
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr20_+_35169897
|
2.783
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr1_+_90098693
|
2.772
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr10_+_75532043
|
2.750
|
NM_173540
|
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase)
|
|
chr17_+_38119225
|
2.748
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr2_-_216300770
|
2.744
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr8_-_145018904
|
2.742
|
NM_201381
|
PLEC
|
plectin
|
|
chr2_-_216300444
|
2.740
|
|
FN1
|
fibronectin 1
|
|
chr1_+_160175146
|
2.740
|
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr20_+_36531498
|
2.724
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr1_-_153044083
|
2.713
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr4_-_2936563
|
2.706
|
NM_001146069
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr17_-_62657997
|
2.687
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr18_+_12948010
|
2.686
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr12_+_58003962
|
2.685
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr22_-_39639967
|
2.684
|
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr20_-_62711160
|
2.681
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr3_+_32433342
|
2.671
|
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr7_-_143059144
|
2.661
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr1_+_90098643
|
2.659
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr12_+_111843743
|
2.644
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr11_+_69455837
|
2.635
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr8_+_25042254
|
2.632
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr1_+_26606443
|
2.630
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr20_+_44746948
|
2.620
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr19_-_19006868
|
2.602
|
NM_021267
NM_198207
NM_001492
|
CERS1
GDF1
|
ceramide synthase 1
growth differentiation factor 1
|
|
chr19_-_19052032
|
2.588
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr18_+_77155966
|
2.581
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_-_45267620
|
2.580
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr6_+_35310317
|
2.576
|
NM_001171818
NM_001171819
NM_001171820
NM_006238
NM_177435
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr1_-_36851517
|
2.572
|
|
STK40
|
serine/threonine kinase 40
|
|
chr9_-_86153048
|
2.571
|
NM_001244959
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr19_+_17858504
|
2.570
|
NM_001161357
NM_001161358
|
FCHO1
|
FCH domain only 1
|
|
chr2_+_173292306
|
2.565
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr1_+_15736387
|
2.557
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr22_-_28315150
|
2.556
|
|
PITPNB
|
phosphatidylinositol transfer protein, beta
|
|
chr6_+_35310336
|
2.548
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr3_-_185542744
|
2.544
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr6_+_35310368
|
2.537
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr10_-_17271955
|
2.534
|
|
|
|
|
chr13_-_20767036
|
2.532
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr19_-_2721337
|
2.531
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr7_-_127670995
|
2.530
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr3_-_27763784
|
2.527
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr1_-_201476248
|
2.525
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr22_-_20255614
|
2.523
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr7_+_143058794
|
2.510
|
|
ZYX
|
zyxin
|
|
chr3_+_32433151
|
2.505
|
NM_138410
NM_181472
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr17_-_46115135
|
2.504
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr16_+_30389632
|
2.497
|
NM_001214906
NM_001214907
|
ZNF48
|
zinc finger protein 48
|
|
chr15_-_40600025
|
2.490
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr2_+_173292081
|
2.486
|
|
ITGA6
|
integrin, alpha 6
|
|
chr22_-_36784055
|
2.464
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr11_+_46403260
|
2.462
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_+_69924407
|
2.460
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr19_+_15218141
|
2.446
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chrX_+_102631408
|
2.445
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr17_-_1083008
|
2.442
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr10_-_62703822
|
2.438
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr1_+_228337414
|
2.438
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr14_-_71275732
|
2.434
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_+_215256559
|
2.417
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr18_+_12308242
|
2.410
|
NM_032525
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr22_-_36784001
|
2.408
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr19_+_12273866
|
2.405
|
NM_003437
|
ZNF136
|
zinc finger protein 136
|
|
chr1_-_6557471
|
2.388
|
NM_001042663
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr3_-_53880403
|
2.387
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr1_-_153067000
|
2.385
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr9_+_125133228
|
2.384
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr19_+_49055421
|
2.382
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr16_+_82660641
|
2.380
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|