|
chr19_+_44556163
|
42.775
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr19_+_37862055
|
20.968
|
NM_032453
|
ZNF527
|
zinc finger protein 527
|
|
chr19_+_54024176
|
20.132
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr19_+_48673936
|
19.339
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr19_-_55628780
|
17.593
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr19_+_54618789
|
16.805
|
NM_015629
|
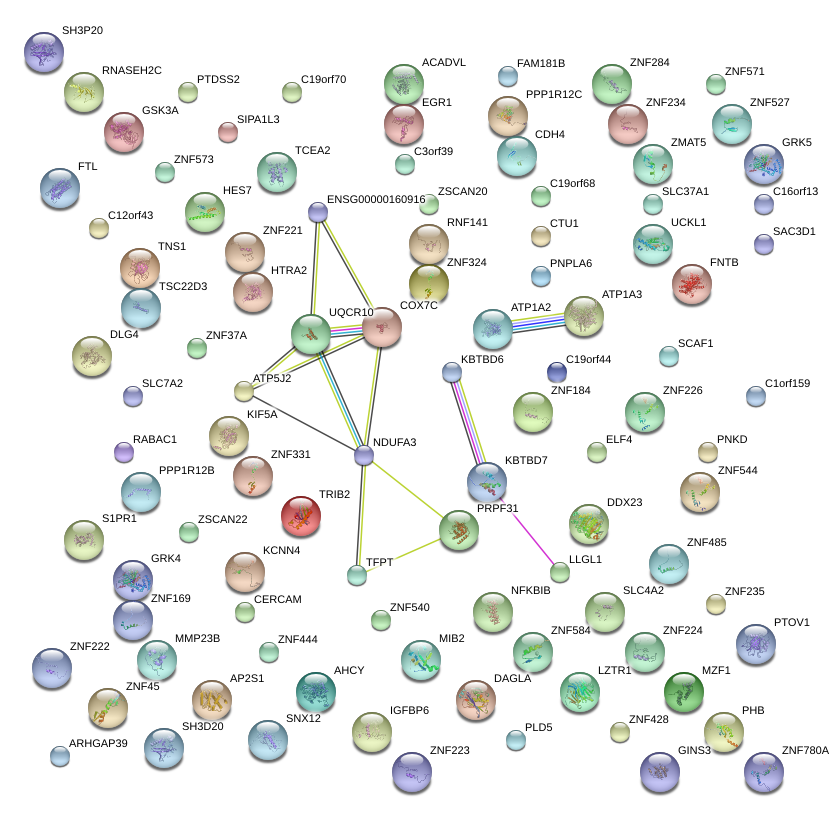
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr19_+_58838343
|
15.596
|
NM_181846
|
ZSCAN22
|
zinc finger and SCAN domain containing 22
|
|
chr19_-_38270199
|
14.838
|
NM_001172689
NM_001172690
NM_001172691
NM_001172692
NM_152360
|
ZNF573
|
zinc finger protein 573
|
|
chr19_-_59084928
|
14.440
|
NM_003422
NM_198055
|
MZF1
|
myeloid zinc finger 1
|
|
chr9_+_97021556
|
14.131
|
NM_194320
|
ZNF169
|
zinc finger protein 169
|
|
chr19_+_58816714
|
13.978
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr19_+_54606141
|
13.364
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr19_+_58816757
|
12.859
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr19_+_49468656
|
12.579
|
|
FTL
|
ferritin, light polypeptide
|
|
chr19_-_59084735
|
12.360
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr19_+_58962954
|
12.242
|
NM_207395
|
ZNF324B
|
zinc finger protein 324B
|
|
chr19_-_59084630
|
12.211
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr19_+_44617510
|
12.148
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr19_-_42746743
|
12.105
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr20_+_59827452
|
12.083
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr11_+_450408
|
11.512
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr19_-_42746678
|
11.420
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr13_-_41768533
|
11.333
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr19_+_38397880
|
11.151
|
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr19_-_54619025
|
11.033
|
NM_013342
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr19_+_56652689
|
10.784
|
|
ZNF444
|
zinc finger protein 444
|
|
chr13_-_41768392
|
10.122
|
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr19_-_42463500
|
10.105
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr20_+_62694009
|
10.052
|
NM_003195
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr19_-_42498320
|
9.947
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr11_+_450260
|
9.934
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr22_+_21336616
|
9.906
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr11_+_450346
|
9.837
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr2_+_74757127
|
9.825
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_+_50145369
|
9.726
|
NM_021228
|
SCAF1
|
SR-related CTD-associated factor 1
|
|
chr22_+_21336508
|
9.617
|
NM_006767
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr19_+_44669214
|
9.612
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr19_+_44507076
|
9.574
|
NM_006300
|
ZNF230
|
zinc finger protein 230
|
|
chr19_-_5680503
|
9.541
|
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr2_+_219187975
|
9.457
|
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr7_+_150755298
|
9.415
|
NM_001199692
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr19_+_44331443
|
9.390
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr20_-_32891075
|
9.250
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr2_+_219187858
|
9.220
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr19_+_56652529
|
9.201
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr19_-_44285012
|
9.180
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr19_-_42463437
|
9.082
|
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr19_-_42746723
|
9.076
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr22_-_30162990
|
8.989
|
|
|
|
|
chr19_+_38397841
|
8.934
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr19_+_58920007
|
8.867
|
NM_173548
|
ZNF584
|
zinc finger protein 584
|
|
chr20_-_62587718
|
8.778
|
NM_017859
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr8_+_17354562
|
8.573
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr13_-_41706587
|
8.562
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr19_-_42746544
|
8.369
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_+_74757243
|
8.359
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr16_-_686292
|
8.346
|
NM_001040160
NM_001040161
NM_001040162
NM_001040165
NM_032366
|
C16orf13
|
chromosome 16 open reading frame 13
|
|
chr17_-_43510204
|
8.293
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr22_+_21336575
|
8.293
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr10_+_38383218
|
8.287
|
NM_001007094
NM_001178101
NM_003421
|
ZNF37A
|
zinc finger protein 37A
|
|
chr2_+_74757116
|
8.175
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr17_-_47492216
|
8.081
|
NM_002634
|
PHB
|
prohibitin
|
|
chr11_+_61447816
|
8.046
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr17_-_7123368
|
8.045
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr19_-_44439410
|
8.023
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr2_-_218867677
|
8.009
|
|
TNS1
|
tensin 1
|
|
chr19_+_44598479
|
7.988
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr19_+_44576266
|
7.910
|
NM_001037813
|
ZNF284
|
zinc finger protein 284
|
|
chr19_+_39390258
|
7.885
|
NM_001243116
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr1_+_1567556
|
7.818
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr17_-_8027396
|
7.804
|
NM_001165967
NM_032580
|
HES7
|
hairy and enhancer of split 7 (Drosophila)
|
|
chr19_-_44123732
|
7.802
|
|
ZNF428
|
zinc finger protein 428
|
|
chr19_-_44285407
|
7.788
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr20_+_62694456
|
7.773
|
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr19_-_44809126
|
7.717
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr17_-_7122901
|
7.651
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr1_-_1051458
|
7.620
|
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr12_-_121454285
|
7.600
|
NM_022895
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr21_+_43919741
|
7.585
|
NM_018964
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr4_+_2965267
|
7.529
|
NM_001004056
NM_001004057
NM_182982
|
GRK4
|
G protein-coupled receptor kinase 4
|
|
chr1_+_101702561
|
7.521
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_-_47492215
|
7.520
|
|
PHB
|
prohibitin
|
|
chr19_-_5680910
|
7.496
|
NM_205767
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr19_-_51611622
|
7.482
|
NM_145232
|
CTU1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe)
|
|
chr11_-_65487910
|
7.437
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr11_+_64808375
|
7.427
|
NM_013299
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr1_+_101702304
|
7.402
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_+_202317798
|
7.387
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr2_+_74757158
|
7.300
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_+_16607181
|
7.288
|
NM_032207
|
C19orf44
|
chromosome 19 open reading frame 44
|
|
chr19_+_44529493
|
7.286
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr11_-_65488142
|
7.284
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr19_-_44617271
|
7.262
|
|
LOC100379224
|
uncharacterized LOC100379224
|
|
chr19_+_50353942
|
7.209
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_+_39390567
|
7.077
|
NM_002503
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr19_-_44124013
|
7.060
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr10_-_43048029
|
7.003
|
|
ZNF37BP
|
zinc finger protein 37B, pseudogene
|
|
chr1_-_242687997
|
6.996
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr19_+_39390618
|
6.979
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr10_+_120967094
|
6.886
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr11_+_64808395
|
6.844
|
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr2_+_74756986
|
6.834
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr3_-_43147470
|
6.789
|
NM_032806
|
C3orf39
|
chromosome 3 open reading frame 39
|
|
chr1_+_33938231
|
6.755
|
NM_145238
|
ZSCAN20
|
zinc finger and SCAN domain containing 20
|
|
chr19_+_38085730
|
6.717
|
NM_001172225
NM_001172226
|
ZNF540
|
zinc finger protein 540
|
|
chr22_-_30162933
|
6.689
|
NM_001003692
NM_019103
|
ZMAT5
|
zinc finger, matrin-type 5
|
|
chr12_-_49245904
|
6.585
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr19_+_58740069
|
6.561
|
NM_014480
|
ZNF544
|
zinc finger protein 544
|
|
chr19_+_7600581
|
6.534
|
NM_001166114
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr19_+_58740232
|
6.493
|
|
ZNF544
|
zinc finger protein 544
|
|
chr22_+_30163351
|
6.441
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr9_+_131182690
|
6.419
|
NM_016174
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr19_-_40596781
|
6.405
|
NM_001010880
NM_001142577
NM_001142578
NM_001142579
|
ZNF780A
|
zinc finger protein 780A
|
|
chr5_+_85913755
|
6.331
|
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr12_+_53491448
|
6.324
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr19_+_54619114
|
6.314
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr19_-_59084089
|
6.312
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr19_+_58978336
|
6.235
|
NM_014347
|
ZNF324
|
zinc finger protein 324
|
|
chr8_-_145911160
|
6.228
|
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr7_-_99063763
|
6.224
|
NM_001198879
NM_001003713
NM_001003714
NM_001039178
NM_004889
|
ATP5J2-PTCD1
ATP5J2
|
ATP5J2-PTCD1 readthrough
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2
|
|
chr17_-_43510279
|
6.201
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chrX_-_106960268
|
6.181
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_+_12856813
|
6.176
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr1_+_1550794
|
6.165
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr19_+_7600632
|
6.164
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr17_+_7123246
|
6.156
|
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr6_-_27440459
|
6.139
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr14_+_65453439
|
6.099
|
NM_002028
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr19_-_47354060
|
6.089
|
NM_004069
NM_021575
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr10_+_44101854
|
6.072
|
NM_145312
|
ZNF485
|
zinc finger protein 485
|
|
chr19_+_44645708
|
6.022
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr16_+_58426297
|
6.020
|
NM_001126129
NM_001126130
NM_022770
|
GINS3
|
GINS complex subunit 3 (Psf3 homolog)
|
|
chr19_+_3185927
|
6.018
|
|
NCLN
|
nicalin
|
|
chr16_+_577833
|
5.961
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr3_+_196729776
|
5.943
|
|
MFI2-AS1
|
MFI2 antisense RNA 1 (non-protein coding)
|
|
chr19_+_3185965
|
5.938
|
|
NCLN
|
nicalin
|
|
chr20_-_62339205
|
5.935
|
|
ARFRP1
|
ADP-ribosylation factor related protein 1
|
|
chr17_-_21117899
|
5.890
|
NM_003876
|
TMEM11
|
transmembrane protein 11
|
|
chr16_-_58718607
|
5.839
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr16_-_3285359
|
5.830
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr22_+_50624353
|
5.804
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr14_+_65453605
|
5.745
|
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr9_-_19102898
|
5.733
|
NM_017645
|
HAUS6
|
HAUS augmin-like complex, subunit 6
|
|
chr19_+_3185860
|
5.704
|
NM_020170
|
NCLN
|
nicalin
|
|
chr19_+_54619149
|
5.697
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr19_+_30156326
|
5.689
|
NM_024310
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr8_-_28747422
|
5.662
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr12_+_56546203
|
5.651
|
NM_001199629
NM_002475
|
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle
|
|
chr19_+_44645728
|
5.645
|
|
ZNF234
|
zinc finger protein 234
|
|
chr1_-_1051735
|
5.631
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr19_+_58790306
|
5.601
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr19_+_58920118
|
5.543
|
|
ZNF584
|
zinc finger protein 584
|
|
chr22_+_50624267
|
5.539
|
|
TRABD
|
TraB domain containing
|
|
chr7_+_150725502
|
5.527
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr7_-_150924220
|
5.516
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr9_-_127533397
|
5.514
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr20_-_62339309
|
5.514
|
NM_001134758
NM_003224
|
ARFRP1
|
ADP-ribosylation factor related protein 1
|
|
chr17_+_900329
|
5.512
|
NM_013337
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast)
|
|
chrX_+_48334400
|
5.498
|
NM_012280
NM_177439
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr22_-_46449972
|
5.467
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr17_+_7123149
|
5.436
|
NM_000018
NM_001033859
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr19_-_1021122
|
5.398
|
NM_001033026
NM_033420
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr19_+_58987751
|
5.352
|
NM_017908
|
ZNF446
|
zinc finger protein 446
|
|
chr12_+_57881853
|
5.330
|
|
MARS
|
methionyl-tRNA synthetase
|
|
chr1_-_26324534
|
5.330
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr12_-_121454210
|
5.285
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr6_+_56911422
|
5.277
|
|
KIAA1586
|
KIAA1586
|
|
chr2_-_86332930
|
5.266
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr19_-_633526
|
5.262
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr12_+_56498347
|
5.262
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr11_-_65314049
|
5.220
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr4_-_2965147
|
5.210
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr16_+_2867227
|
5.206
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr19_+_35225095
|
5.197
|
|
ZNF181
|
zinc finger protein 181
|
|
chr19_+_40697516
|
5.193
|
NM_002446
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr2_+_217277136
|
5.164
|
NM_014140
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1
|
|
chr12_+_57881735
|
5.147
|
NM_004990
|
MARS
|
methionyl-tRNA synthetase
|
|
chr19_+_57106622
|
5.145
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr17_-_21117647
|
5.128
|
|
TMEM11
|
transmembrane protein 11
|
|
chr19_-_38085646
|
5.048
|
NM_016536
|
ZNF571
|
zinc finger protein 571
|
|
chr12_+_56498414
|
5.047
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr2_+_130939781
|
5.041
|
|
MZT2B
|
mitotic spindle organizing protein 2B
|
|
chr12_+_56498365
|
4.998
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr14_+_55034633
|
4.981
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_10532603
|
4.976
|
NM_004401
NM_213566
|
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide
|
|
chr19_-_1020995
|
4.969
|
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr19_+_46010711
|
4.953
|
|
VASP
|
vasodilator-stimulated phosphoprotein
|
|
chr19_-_57988891
|
4.920
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr12_+_56498334
|
4.899
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr6_+_42531736
|
4.889
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr10_-_13390189
|
4.878
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr7_+_112090463
|
4.865
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr16_-_31106125
|
4.863
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr19_+_54371150
|
4.860
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_+_156024516
|
4.858
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr2_+_201450730
|
4.854
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr9_-_19102648
|
4.844
|
|
HAUS6
|
HAUS augmin-like complex, subunit 6
|
|
chr9_+_15422875
|
4.843
|
|
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa
|
|
chr12_+_120740123
|
4.837
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr19_-_39390168
|
4.797
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|