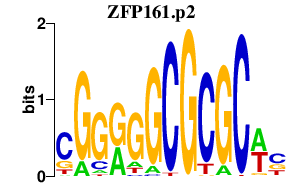
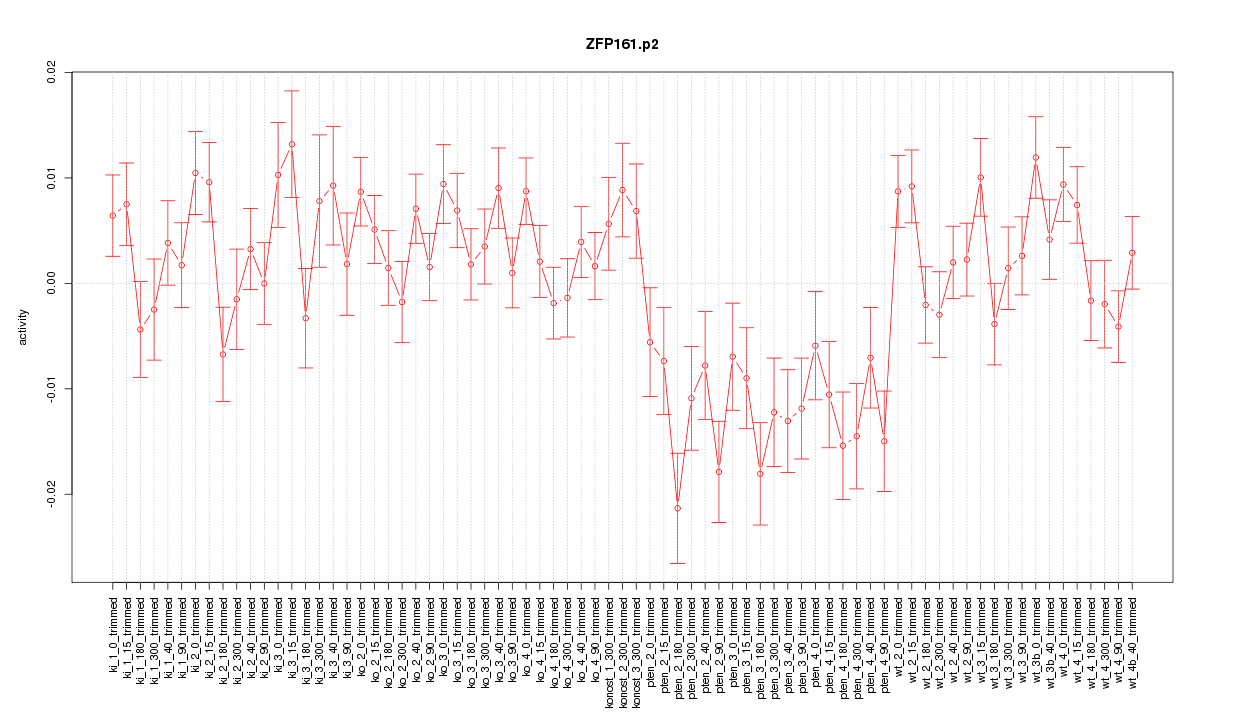
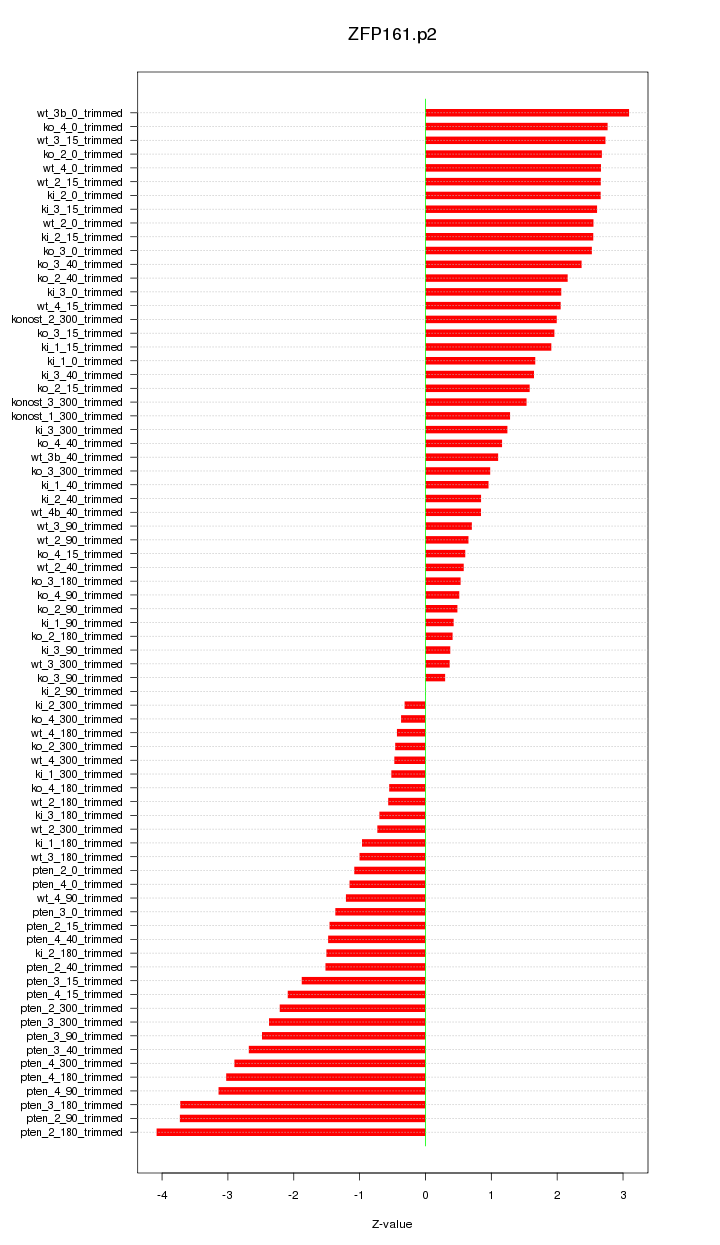
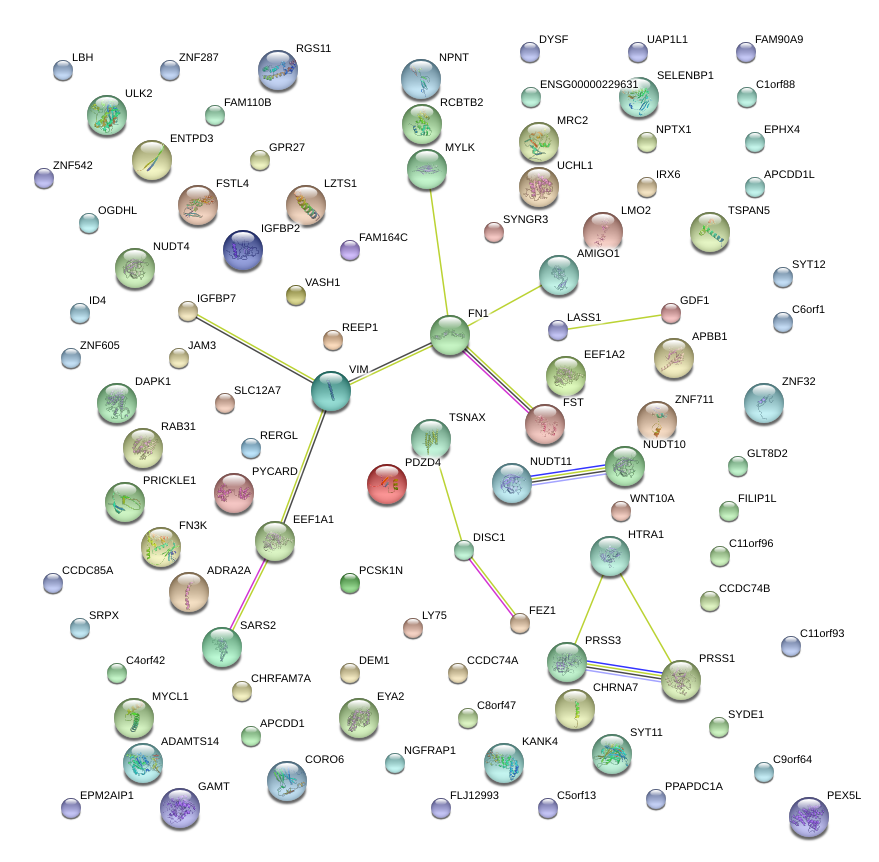
|
chr17_-_19770988
|
28.414
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr2_+_30454396
|
26.889
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr11_-_125366048
|
20.046
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr17_+_60705051
|
19.945
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr16_-_325867
|
19.893
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr17_+_60705042
|
19.214
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr12_-_133532847
|
19.070
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr17_+_60704747
|
18.833
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr19_+_15218141
|
17.817
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr1_+_92495484
|
17.202
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr16_+_55358470
|
15.898
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr1_-_110052285
|
15.364
|
NM_020703
|
AMIGO1
|
adhesion molecule with Ig-like domain 1
|
|
chr11_-_6440288
|
14.997
|
NM_001164
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr2_+_71693817
|
14.861
|
NM_001130455
NM_001130982
NM_001130983
NM_001130984
NM_001130985
NM_001130986
NM_001130987
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr4_+_106816593
|
14.340
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr9_-_86571533
|
13.815
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr1_-_40367539
|
13.514
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chrX_+_102631408
|
13.408
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr9_-_86571652
|
13.130
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr11_-_6440631
|
11.838
|
NM_145689
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr3_-_99594915
|
11.113
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr16_+_2039945
|
10.897
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chrX_+_84498993
|
10.740
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr17_-_16472463
|
10.706
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chrX_-_153095996
|
10.684
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr1_-_62785067
|
10.581
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr11_+_43963809
|
10.501
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr1_-_1535446
|
10.443
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chrX_-_153095810
|
10.309
|
|
PDZD4
|
PDZ domain containing 4
|
|
chrX_+_102631267
|
10.271
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr15_+_32322690
|
10.168
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr14_+_75536298
|
10.082
|
NM_001042430
NM_024643
|
FAM164C
|
family with sequence similarity 164, member C
|
|
chr4_+_41258941
|
10.048
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr19_-_19006868
|
9.960
|
NM_021267
NM_198207
NM_001492
|
CERS1
GDF1
|
ceramide synthase 1
growth differentiation factor 1
|
|
chrX_-_51239295
|
9.842
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr4_-_99579540
|
9.574
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr2_-_86564632
|
9.571
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr1_-_62784971
|
9.455
|
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chrX_-_38079980
|
9.432
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr8_+_58906968
|
9.426
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr19_-_1401474
|
9.360
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr18_+_10454620
|
9.310
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr20_-_62130390
|
9.184
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr4_+_106816627
|
9.163
|
|
NPNT
|
nephronectin
|
|
chr17_-_27949814
|
9.161
|
|
CORO6
|
coronin 6
|
|
chr11_+_66790815
|
9.030
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr11_-_33890931
|
8.836
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr2_+_217498082
|
8.813
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr19_+_56879687
|
8.668
|
|
ZNF542
|
zinc finger protein 542
|
|
chr3_-_179754516
|
8.159
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr2_-_216300444
|
8.014
|
|
FN1
|
fibronectin 1
|
|
chr4_-_57976550
|
7.949
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr17_+_80693398
|
7.929
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr5_+_52776479
|
7.853
|
|
FST
|
follistatin
|
|
chrX_-_51239426
|
7.825
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr10_-_44144266
|
7.818
|
NM_006973
|
ZNF32
|
zinc finger protein 32
|
|
chr18_+_9708322
|
7.814
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr9_+_33750463
|
7.797
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr10_-_44144151
|
7.771
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr6_+_19837599
|
7.700
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr8_+_72756218
|
7.607
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr20_+_56285213
|
7.515
|
|
|
|
|
chr10_-_126138549
|
7.496
|
NM_001146340
|
NKX1-2
|
NK1 homeobox 2
|
|
chr10_-_50970321
|
7.390
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chrX_-_48693934
|
7.363
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr4_-_83483099
|
7.358
|
NM_001080506
|
TMEM150C
|
transmembrane protein 150C
|
|
chr16_-_31214054
|
7.346
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chrX_+_84499037
|
7.342
|
|
ZNF711
|
zinc finger protein 711
|
|
chr14_+_77228132
|
7.320
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr17_-_78450247
|
7.281
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr2_-_160761192
|
7.246
|
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr13_-_49107250
|
7.191
|
NM_001268
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr5_-_111093251
|
7.165
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_17270469
|
7.153
|
|
VIM
|
vimentin
|
|
chr5_-_132948222
|
7.108
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr1_+_231762520
|
7.091
|
NM_001012957
NM_001012958
NM_001012959
NM_001164537
NM_001164538
NM_001164539
NM_001164540
NM_001164541
NM_001164542
NM_001164544
NM_001164545
NM_001164546
NM_001164547
NM_001164548
NM_001164549
NM_001164550
NM_001164551
NM_001164552
NM_001164553
NM_001164554
NM_001164555
NM_001164556
NM_018662
|
DISC1
|
disrupted in schizophrenia 1
|
|
chr1_+_40974430
|
7.078
|
NM_022774
|
DEM1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr3_+_71803200
|
7.071
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr14_+_77228940
|
6.995
|
|
VASH1
|
vasohibin 1
|
|
chr10_+_72432557
|
6.923
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr18_+_9708187
|
6.920
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr18_+_9708269
|
6.866
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_-_1112106
|
6.844
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr4_+_41258909
|
6.785
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr10_+_122216465
|
6.733
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr20_+_45523247
|
6.640
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr20_-_57089845
|
6.569
|
NM_153360
|
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like
|
|
chr3_+_48885369
|
6.563
|
|
|
|
|
chr11_+_111169965
|
6.537
|
NM_001136105
|
C11orf93
|
chromosome 11 open reading frame 93
|
|
chr2_+_219745243
|
6.526
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr9_-_86571631
|
6.486
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr12_-_42983412
|
6.469
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr8_+_99076749
|
6.413
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr16_-_31214003
|
6.383
|
|
PYCARD
|
PYD and CARD domain containing
|
|
chr9_+_139972195
|
6.341
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr11_+_133938965
|
6.238
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr9_+_90112419
|
6.222
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr9_+_139971938
|
6.176
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr11_+_133938819
|
6.144
|
NM_001205329
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr4_+_1242942
|
6.095
|
|
C4orf42
|
chromosome 4 open reading frame 42
|
|
chr1_+_111889194
|
6.087
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr3_+_40428672
|
6.073
|
NM_001248
|
ENTPD3
|
ectonucleoside triphosphate diphosphohydrolase 3
|
|
chr10_+_124220990
|
6.070
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chrX_-_153095746
|
6.054
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr3_-_123603144
|
6.025
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr20_-_44563684
|
6.013
|
|
FLJ40606
|
uncharacterized LOC643549
|
|
chr2_+_56411123
|
5.972
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr5_+_52776263
|
5.971
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr2_+_132285469
|
5.967
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr5_-_168727715
|
5.938
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr18_-_45935627
|
5.895
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr13_-_49107185
|
5.864
|
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr18_+_10454542
|
5.854
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr10_-_62761197
|
5.838
|
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr10_-_98592186
|
5.794
|
|
|
|
|
chr12_-_99038705
|
5.775
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr19_-_18717467
|
5.753
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr2_+_37571752
|
5.740
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr8_-_17658385
|
5.712
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr14_-_92413738
|
5.708
|
|
FBLN5
|
fibulin 5
|
|
chr19_-_19007395
|
5.672
|
|
CERS1
|
ceramide synthase 1
|
|
chr11_-_33891258
|
5.629
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_-_25038501
|
5.598
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr3_-_53880403
|
5.592
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr19_-_37329258
|
5.587
|
NM_206894
|
ZNF790
|
zinc finger protein 790
|
|
chr19_+_7580987
|
5.554
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr8_+_72756335
|
5.538
|
|
|
|
|
chr1_-_85358851
|
5.531
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr5_+_121647773
|
5.480
|
NM_001242935
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chrX_-_64254592
|
5.474
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr11_-_108368918
|
5.460
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr2_-_45236521
|
5.458
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr7_+_45614081
|
5.407
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr2_-_225907312
|
5.346
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr4_+_41258895
|
5.345
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr3_-_38691118
|
5.343
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_+_154301219
|
5.329
|
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr15_+_99645152
|
5.279
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr16_+_28303622
|
5.196
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr6_+_13925097
|
5.190
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chrX_-_17879355
|
5.181
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr18_+_10454625
|
5.055
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr3_-_126076057
|
5.036
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr14_+_77227783
|
5.006
|
|
|
|
|
chr6_-_36355342
|
4.979
|
NM_001207038
NM_001207035
NM_001207036
NM_001207037
NM_001207039
NM_001207040
NM_001207041
NM_016135
|
ETV7
|
ets variant 7
|
|
chr7_+_89840999
|
4.939
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr7_+_2559396
|
4.926
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr12_-_99038401
|
4.922
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr17_-_46655544
|
4.857
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr14_+_53019865
|
4.848
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr20_+_45523505
|
4.829
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr3_-_160822625
|
4.824
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr5_-_136834887
|
4.823
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr7_+_45614179
|
4.766
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr22_+_42394728
|
4.757
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr5_+_137774632
|
4.745
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr6_-_134496833
|
4.725
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_-_92414004
|
4.703
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr9_-_99801365
|
4.702
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr2_-_216300770
|
4.690
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr19_+_33622997
|
4.662
|
NM_173479
|
WDR88
|
WD repeat domain 88
|
|
chr14_-_105444693
|
4.651
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr14_-_65879245
|
4.639
|
|
LOC645431
|
uncharacterized LOC645431
|
|
chr11_-_57335080
|
4.636
|
NM_004223
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr11_+_129245834
|
4.619
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr16_-_30546145
|
4.596
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr6_-_89827605
|
4.594
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr6_-_170599568
|
4.587
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr17_+_79885567
|
4.573
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr14_+_74058409
|
4.554
|
NM_152331
|
ACOT4
|
acyl-CoA thioesterase 4
|
|
chr7_+_73082157
|
4.540
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr5_-_111093792
|
4.524
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_50712671
|
4.511
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr1_-_24513737
|
4.490
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr7_+_121513158
|
4.477
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr20_+_45523523
|
4.464
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr10_+_95753684
|
4.461
|
NM_016341
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr2_-_160761266
|
4.443
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr19_-_32896441
|
4.421
|
|
LOC400684
|
uncharacterized LOC400684
|
|
chr3_-_55521330
|
4.416
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr2_-_228028765
|
4.378
|
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr5_-_134914447
|
4.315
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr6_-_111136407
|
4.285
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr22_-_28197469
|
4.279
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr9_+_129376721
|
4.277
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr5_+_172483285
|
4.248
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr16_-_28303366
|
4.221
|
|
|
|
|
chr16_-_54962707
|
4.192
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr11_+_118401755
|
4.191
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr12_+_133657478
|
4.190
|
|
ZNF140
|
zinc finger protein 140
|
|
chr3_+_186648515
|
4.186
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr16_-_1968230
|
4.183
|
NM_001009606
|
HS3ST6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6
|
|
chr9_+_140033588
|
4.177
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr6_+_13925423
|
4.174
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr6_-_111136330
|
4.155
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr2_+_150187028
|
4.109
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr11_-_33891361
|
4.103
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr7_+_29234325
|
4.098
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr1_+_204797774
|
4.094
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr14_-_105886075
|
4.075
|
|
LOC100507437
|
uncharacterized LOC100507437
|