|
chr17_+_54671059
|
5.345
|
NM_005450
|
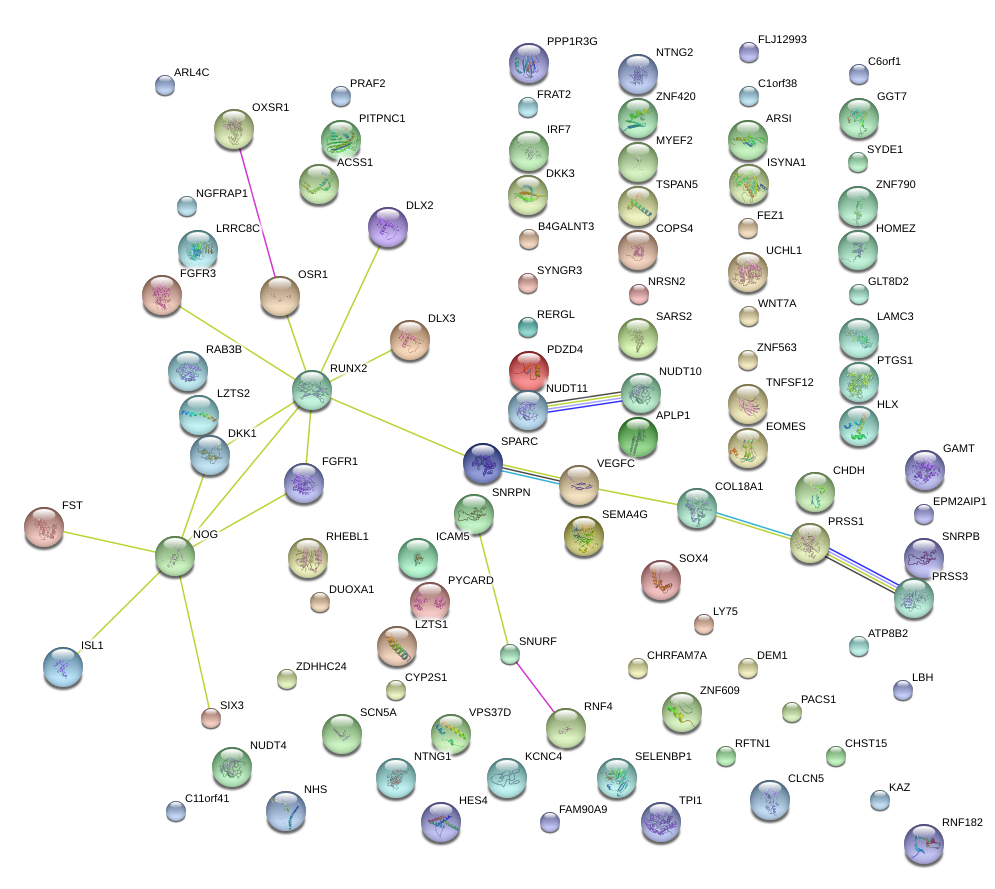
NOG
|
noggin
|
|
chrX_+_102632108
|
4.743
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chrX_-_153095996
|
4.432
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr3_-_53880403
|
4.162
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr16_-_31214054
|
4.003
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chr15_+_25200069
|
3.741
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr1_+_28199053
|
3.719
|
NM_001039477
NM_001105556
NM_004848
|
C1orf38
|
chromosome 1 open reading frame 38
|
|
chr4_+_41258941
|
3.708
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr19_+_15218141
|
3.587
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr5_-_149682447
|
3.521
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr16_-_31214003
|
3.269
|
|
PYCARD
|
PYD and CARD domain containing
|
|
chr20_-_33460558
|
3.080
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr1_+_77165438
|
2.983
|
|
TPI1
|
triosephosphate isomerase 1
|
|
chr7_+_73082157
|
2.922
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chrX_-_51239295
|
2.920
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr20_+_327725
|
2.890
|
|
NRSN2
|
neurensin 2
|
|
chr4_-_99579540
|
2.847
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr19_+_36359346
|
2.845
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr10_-_99094264
|
2.823
|
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr11_-_125366048
|
2.792
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr4_+_41258909
|
2.677
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr17_+_7452374
|
2.628
|
NM_172089
NM_003809
|
TNFSF12-TNFSF13
TNFSF12
|
TNFSF12-TNFSF13 readthrough
tumor necrosis factor (ligand) superfamily, member 12
|
|
chr1_+_40974430
|
2.558
|
NM_022774
|
DEM1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr2_+_30454396
|
2.547
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr12_+_569542
|
2.533
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr6_+_21593963
|
2.491
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr11_-_12030882
|
2.441
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr5_-_151066492
|
2.413
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr19_-_1401474
|
2.402
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr1_-_52456292
|
2.387
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr4_-_177713664
|
2.385
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr1_+_107682628
|
2.335
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr19_+_41699076
|
2.335
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr20_-_25038501
|
2.302
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chrX_-_51239426
|
2.269
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr11_-_12030128
|
2.243
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr6_+_5085719
|
2.134
|
NM_001145115
|
PPP1R3G
|
protein phosphatase 1, regulatory subunit 3G
|
|
chr5_+_52776479
|
2.133
|
|
FST
|
follistatin
|
|
chr4_+_41258895
|
2.132
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr19_+_37569307
|
2.096
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|
|
chr1_-_935352
|
2.094
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr15_-_30685562
|
2.090
|
NM_139320
NM_148911
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion
|
|
chr4_-_83483099
|
2.077
|
NM_001080506
|
TMEM150C
|
transmembrane protein 150C
|
|
chr3_-_38691118
|
2.075
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_+_90098624
|
2.061
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr5_-_151066445
|
2.006
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr19_-_12444435
|
1.998
|
NM_145276
|
ZNF563
|
zinc finger protein 563
|
|
chr9_+_125133228
|
1.985
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr11_-_615920
|
1.976
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr4_+_1795004
|
1.949
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr2_-_160761266
|
1.944
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr19_-_18548788
|
1.912
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr1_+_154301219
|
1.909
|
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr10_+_54074037
|
1.906
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr5_-_141257827
|
1.871
|
|
|
|
|
chr15_-_45422056
|
1.841
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr2_-_235405692
|
1.815
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr11_-_12030622
|
1.814
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr19_+_41699140
|
1.808
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr9_+_125132823
|
1.806
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr1_+_221052735
|
1.795
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr10_+_22540986
|
1.789
|
|
LOC100130992
|
uncharacterized LOC100130992
|
|
chr1_+_221052845
|
1.780
|
|
HLX
|
H2.0-like homeobox
|
|
chr19_-_37329258
|
1.776
|
NM_206894
|
ZNF790
|
zinc finger protein 790
|
|
chrX_-_48931645
|
1.771
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr5_+_50679239
|
1.748
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr17_+_65373574
|
1.743
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr11_-_615998
|
1.739
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr8_-_38325524
|
1.714
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr11_+_33563772
|
1.703
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr15_+_64752778
|
1.695
|
|
ZNF609
|
zinc finger protein 609
|
|
chr12_-_49463729
|
1.689
|
NM_144593
|
RHEBL1
|
Ras homolog enriched in brain like 1
|
|
chrX_+_17393537
|
1.689
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr21_+_46825001
|
1.688
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr17_+_65373362
|
1.686
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr11_-_615649
|
1.677
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr9_+_33750463
|
1.667
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr7_+_73082320
|
1.650
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr6_-_43478423
|
1.619
|
NM_001012974
|
LRRC73
|
leucine rich repeat containing 73
|
|
chr3_+_159943628
|
1.580
|
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr15_-_48470453
|
1.580
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr1_+_221053000
|
1.565
|
|
HLX
|
H2.0-like homeobox
|
|
chr9_+_125137600
|
1.564
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr11_-_66313474
|
1.543
|
|
ZDHHC24
|
zinc finger, DHHC-type containing 24
|
|
chr8_+_72756218
|
1.534
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr10_+_102729249
|
1.501
|
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr3_-_16555128
|
1.498
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chrX_+_49687212
|
1.489
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr9_+_133884503
|
1.454
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr2_+_45169036
|
1.439
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr19_-_18548971
|
1.434
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr3_-_27763784
|
1.434
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr5_+_50678957
|
1.425
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr16_+_2039945
|
1.423
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr10_-_125853102
|
1.420
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr1_+_110753313
|
1.418
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr10_+_102759607
|
1.418
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr3_-_13921617
|
1.411
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr2_-_19558371
|
1.393
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr17_-_48072566
|
1.379
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr6_+_45389818
|
1.375
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chr6_+_13925097
|
1.371
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chr2_+_88472657
|
1.356
|
NM_018271
|
THNSL2
|
threonine synthase-like 2 (S. cerevisiae)
|
|
chr15_+_80696569
|
1.341
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr19_+_4304590
|
1.340
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr3_+_19189969
|
1.327
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr3_-_16554992
|
1.319
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr15_+_74218962
|
1.317
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_2680349
|
1.313
|
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr13_+_51796363
|
1.289
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr16_+_67686813
|
1.286
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr14_+_104605441
|
1.286
|
|
KIF26A
|
kinesin family member 26A
|
|
chr3_+_159943252
|
1.285
|
NM_001168214
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr1_+_226736603
|
1.277
|
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr10_+_99079017
|
1.275
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr10_+_81892497
|
1.275
|
|
PLAC9
|
placenta-specific 9
|
|
chr17_+_79885567
|
1.270
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr9_-_21559696
|
1.268
|
|
MIR31HG
|
MIR31 host gene (non-protein coding)
|
|
chr5_+_52776408
|
1.267
|
|
FST
|
follistatin
|
|
chr8_-_82024277
|
1.267
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr15_-_45480160
|
1.265
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr3_-_27525878
|
1.262
|
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr19_+_33793743
|
1.255
|
|
LOC80054
|
uncharacterized LOC80054
|
|
chr1_+_90098693
|
1.254
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_-_149982528
|
1.251
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr16_+_3096681
|
1.248
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr20_-_25013307
|
1.242
|
NM_001252676
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr22_-_20792107
|
1.241
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr17_+_65373923
|
1.235
|
NM_012417
NM_181671
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr21_+_47518012
|
1.234
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr20_-_3218834
|
1.229
|
NM_001174090
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr17_-_39743085
|
1.225
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr1_+_90098643
|
1.223
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chrX_+_17393806
|
1.212
|
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr9_+_3526047
|
1.204
|
|
|
|
|
chr6_+_161412826
|
1.201
|
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr11_-_66313509
|
1.192
|
NM_207340
|
ZDHHC24
|
zinc finger, DHHC-type containing 24
|
|
chr19_-_19843766
|
1.188
|
NM_021030
|
ZNF14
|
zinc finger protein 14
|
|
chr17_-_62657997
|
1.187
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr3_+_183415540
|
1.185
|
NM_018023
|
YEATS2
|
YEATS domain containing 2
|
|
chr19_-_19007395
|
1.185
|
|
CERS1
|
ceramide synthase 1
|
|
chr2_-_25536762
|
1.179
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr6_+_151646634
|
1.172
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr16_+_22825806
|
1.169
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr10_+_81892257
|
1.167
|
NM_001012973
|
PLAC9
|
placenta-specific 9
|
|
chr19_+_34972866
|
1.160
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr10_+_102756829
|
1.149
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr7_+_73703704
|
1.147
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr2_+_217498082
|
1.146
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr10_+_101292689
|
1.144
|
NM_145285
|
NKX2-3
|
NK2 homeobox 3
|
|
chr10_-_99094427
|
1.144
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr4_+_42399855
|
1.140
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr2_+_217498205
|
1.138
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr11_-_12030838
|
1.123
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_+_66790189
|
1.122
|
NM_001177880
|
SYT12
|
synaptotagmin XII
|
|
chr19_-_7990974
|
1.111
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr11_-_12030539
|
1.107
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chrX_+_51075667
|
1.107
|
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr10_-_43762366
|
1.104
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr19_+_45312294
|
1.101
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr1_-_201342318
|
1.098
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chrX_+_55478537
|
1.097
|
NM_014061
|
MAGEH1
|
melanoma antigen family H, 1
|
|
chr12_-_133263988
|
1.097
|
|
POLE
|
polymerase (DNA directed), epsilon
|
|
chr5_-_141257943
|
1.095
|
NM_002587
NM_032420
|
PCDH1
|
protocadherin 1
|
|
chr16_+_30935895
|
1.094
|
NM_001099784
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr19_-_41934634
|
1.084
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chrX_-_153095841
|
1.084
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr17_-_40346427
|
1.082
|
NM_001142622
NM_001142623
NM_032484
|
GHDC
|
GH3 domain containing
|
|
chr5_-_176057324
|
1.082
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr10_-_62761197
|
1.079
|
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr14_-_37051785
|
1.077
|
NM_014360
|
NKX2-8
|
NK2 homeobox 8
|
|
chr22_-_37499456
|
1.074
|
NM_153609
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr12_+_111471827
|
1.068
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr16_+_30194730
|
1.064
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr1_-_223537400
|
1.049
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr17_-_73874056
|
1.044
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr17_+_38599651
|
1.034
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr16_+_77233348
|
1.033
|
NM_001129979
|
SYCE1L
|
synaptonemal complex central element protein 1-like
|
|
chr1_+_45965855
|
1.031
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr15_-_75248925
|
1.023
|
|
RPP25
|
ribonuclease P/MRP 25kDa subunit
|
|
chr20_+_35169876
|
1.023
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr7_-_143059696
|
1.019
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr13_+_28494136
|
1.017
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr9_+_125133322
|
1.013
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr19_-_40732563
|
1.011
|
NM_024877
|
CNTD2
|
cyclin N-terminal domain containing 2
|
|
chr10_-_116164514
|
1.011
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr10_-_105614952
|
1.007
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_+_46972667
|
1.007
|
NM_147192
NM_172225
|
DMBX1
|
diencephalon/mesencephalon homeobox 1
|
|
chr8_-_38325241
|
1.003
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr14_-_61190458
|
1.001
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chrX_+_118892544
|
0.987
|
NM_001105576
|
ANKRD58
|
ankyrin repeat domain 58
|
|
chr13_+_42031525
|
0.987
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr19_-_11529948
|
0.984
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr22_-_20792047
|
0.983
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr22_+_51039178
|
0.977
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chrX_-_54521866
|
0.975
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr5_+_86564727
|
0.975
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr5_+_86564226
|
0.971
|
|
|
|
|
chr2_-_27531113
|
0.969
|
NM_003353
|
UCN
|
urocortin
|
|
chr1_+_180123975
|
0.969
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|