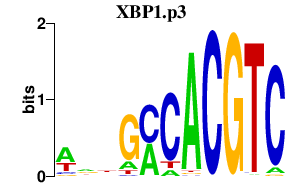
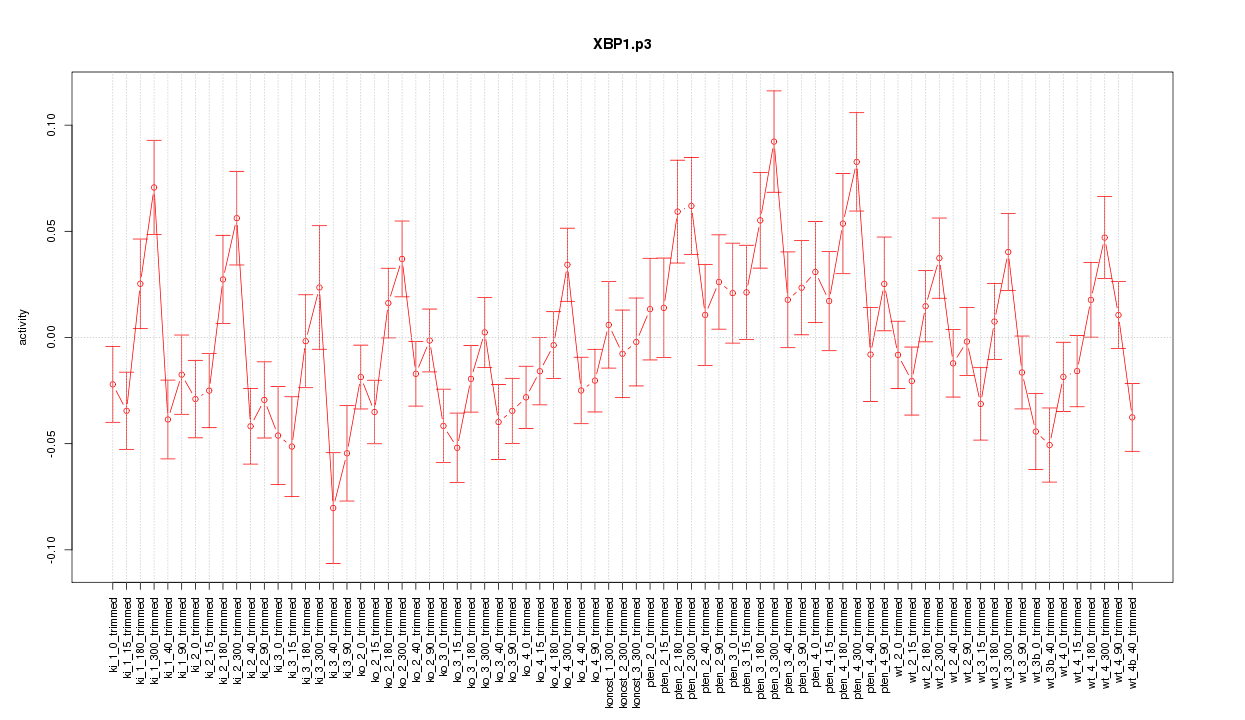
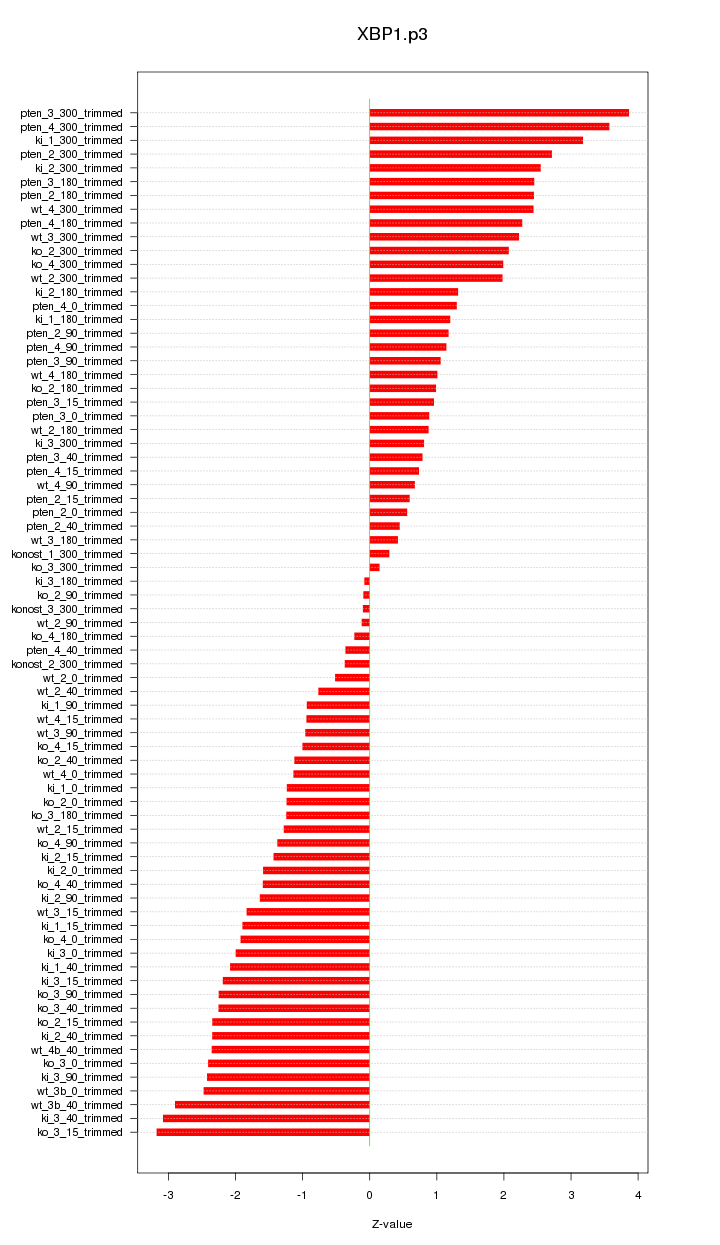
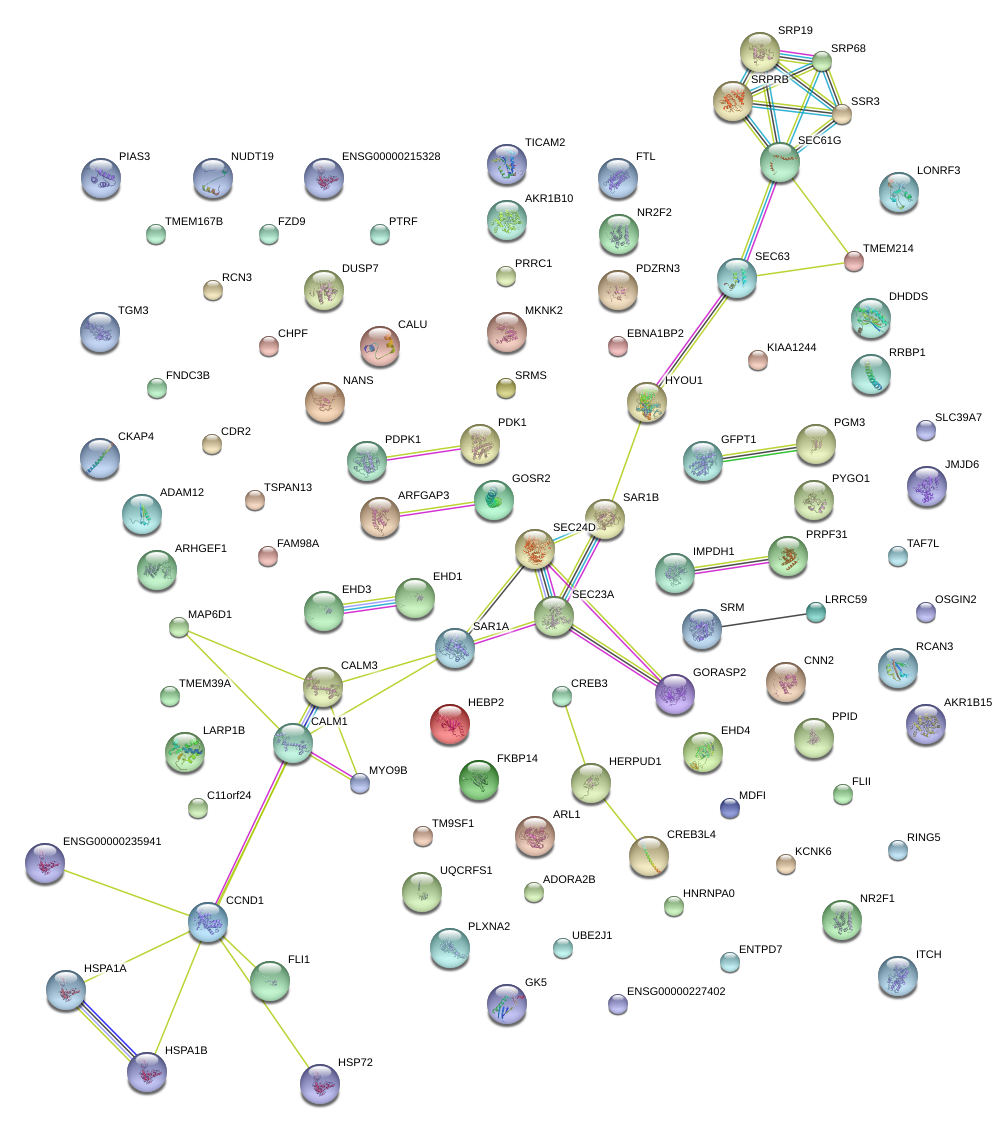
|
chr19_+_49468587
|
10.914
|
|
FTL
|
ferritin, light polypeptide
|
|
chr19_+_49468557
|
10.826
|
NM_000146
|
FTL
|
ferritin, light polypeptide
|
|
chr11_-_2018703
|
8.679
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_+_49468656
|
8.612
|
|
FTL
|
ferritin, light polypeptide
|
|
chr3_-_183543295
|
7.601
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr4_-_119757183
|
7.313
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr1_+_26758772
|
6.970
|
NM_001243564
NM_001243565
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr11_-_118927872
|
6.930
|
NM_006389
NM_001130991
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr7_-_30066124
|
6.730
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr11_-_118927851
|
6.564
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr19_+_47104490
|
6.166
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr19_+_47104559
|
6.049
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr11_-_118927833
|
6.038
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr11_-_118927815
|
5.911
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr11_-_118927756
|
5.877
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr4_-_119757288
|
5.829
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr2_-_33824294
|
5.517
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr5_+_92920592
|
5.397
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_43736600
|
5.376
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr19_+_47104389
|
5.350
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr3_+_133524636
|
4.901
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr3_-_52090090
|
4.842
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr3_+_133524671
|
4.769
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr19_+_42388445
|
4.661
|
NM_199002
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr6_+_41606193
|
4.453
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr9_+_100818941
|
4.430
|
NM_018946
|
NANS
|
N-acetylneuraminic acid synthase
|
|
chr3_-_73673978
|
4.412
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr2_-_69614321
|
4.356
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr6_+_138483024
|
4.262
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr19_+_38810540
|
4.145
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr10_-_128077072
|
4.054
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr7_-_128045995
|
4.042
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr4_+_128982404
|
3.982
|
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr11_-_64646053
|
3.977
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr7_+_72848108
|
3.951
|
NM_003508
|
FZD9
|
frizzled family receptor 9
|
|
chr20_-_17662828
|
3.870
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr16_-_22385596
|
3.838
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr20_-_17662704
|
3.810
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr4_+_128982502
|
3.659
|
NM_018078
NM_032239
NM_178043
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr11_+_69455938
|
3.635
|
|
CCND1
|
cyclin D1
|
|
chr12_-_106641675
|
3.609
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr19_+_33182689
|
3.506
|
NM_001105570
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19
|
|
chr4_+_128982483
|
3.487
|
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr1_-_11120076
|
3.474
|
|
SRM
|
spermidine synthase
|
|
chr5_+_112196976
|
3.420
|
|
SRP19
|
signal recognition particle 19kDa
|
|
chr2_+_171785624
|
3.396
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr5_+_112196884
|
3.361
|
NM_001204193
NM_001204194
NM_001204196
NM_001204199
NM_003135
|
SRP19
|
signal recognition particle 19kDa
|
|
chr11_-_2019070
|
3.359
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_+_118108575
|
3.346
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr20_+_32951054
|
3.344
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr5_-_114938060
|
3.279
|
NM_021649
|
TICAM2
|
toll-like receptor adaptor molecule 2
|
|
chr6_-_108279320
|
3.225
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr7_+_16793320
|
3.173
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr19_-_29704107
|
3.169
|
NM_006003
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr19_+_38810520
|
3.138
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr4_-_159644439
|
3.132
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr1_+_109633373
|
3.114
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr2_+_171785780
|
3.107
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr1_+_145576011
|
3.098
|
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr15_-_55881049
|
3.077
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr6_-_108279212
|
3.075
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr11_-_68039327
|
3.065
|
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr19_-_29704021
|
3.063
|
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr19_+_54618789
|
3.028
|
NM_015629
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr3_-_156272868
|
2.955
|
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr10_-_128077011
|
2.951
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr6_-_90062610
|
2.905
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr17_-_48474864
|
2.893
|
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr5_+_126853308
|
2.865
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr6_-_83902921
|
2.851
|
NM_001199918
NM_001199919
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr1_+_145575987
|
2.800
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr6_-_108279307
|
2.780
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr11_+_128563772
|
2.748
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_-_119182423
|
2.699
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr3_-_156272909
|
2.640
|
NM_007107
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr3_+_171758289
|
2.626
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr7_-_54826864
|
2.569
|
|
SEC61G
|
Sec61 gamma subunit
|
|
chr7_+_128379400
|
2.569
|
|
CALU
|
calumenin
|
|
chr10_-_71930185
|
2.562
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr17_+_15848230
|
2.562
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr6_-_108279409
|
2.552
|
NM_007214
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr11_-_2018334
|
2.547
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_-_11120001
|
2.501
|
|
SRM
|
spermidine synthase
|
|
chr1_-_11120047
|
2.490
|
|
SRM
|
spermidine synthase
|
|
chr22_-_43253186
|
2.487
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr6_+_33168602
|
2.487
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr6_-_108279366
|
2.475
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr19_+_17186569
|
2.468
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr6_-_90062450
|
2.460
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr3_-_141944441
|
2.447
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr7_+_128379428
|
2.391
|
|
CALU
|
calumenin
|
|
chr6_+_31783530
|
2.391
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr6_-_108279378
|
2.367
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr7_+_128379345
|
2.363
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr2_-_220408264
|
2.351
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr10_+_101419169
|
2.338
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr7_+_128379452
|
2.334
|
|
CALU
|
calumenin
|
|
chr1_-_11120081
|
2.332
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr14_-_39572414
|
2.326
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr6_-_90062439
|
2.323
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr17_-_74068583
|
2.310
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr6_-_90062349
|
2.273
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr17_-_74068582
|
2.269
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr14_-_24664758
|
2.267
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr2_+_27255823
|
2.236
|
|
TMEM214
|
transmembrane protein 214
|
|
chr19_-_2038937
|
2.223
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr2_+_27255837
|
2.215
|
|
TMEM214
|
transmembrane protein 214
|
|
chr2_+_173420776
|
2.214
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr19_+_50031232
|
2.212
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr7_+_128379395
|
2.204
|
|
CALU
|
calumenin
|
|
chr17_+_45000493
|
2.201
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr9_+_35732207
|
2.199
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr11_-_68039446
|
2.180
|
NM_022338
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr2_+_27255745
|
2.177
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr17_-_74068522
|
2.174
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr17_-_74068597
|
2.166
|
NM_014230
|
SRP68
|
signal recognition particle 68kDa
|
|
chr17_-_40575117
|
2.156
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr17_-_74068576
|
2.155
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr9_+_35732498
|
2.138
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr19_+_1026582
|
2.129
|
|
CNN2
|
calponin 2
|
|
chr12_-_101801499
|
2.121
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr17_-_74722666
|
2.101
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr15_-_42264749
|
2.091
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr22_-_43253304
|
2.077
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr1_-_208417635
|
2.075
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr3_-_141944296
|
2.074
|
|
GK5
|
glycerol kinase 5 (putative)
|
|
chrX_+_118108595
|
2.068
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr19_+_1026624
|
2.067
|
|
CNN2
|
calponin 2
|
|
chr5_-_137088952
|
2.050
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr16_+_56966025
|
2.047
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr19_+_38810461
|
2.015
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_+_38810751
|
2.013
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr2_+_173420794
|
2.010
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr15_-_60690115
|
1.992
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr2_-_88926863
|
1.986
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr19_+_45394522
|
1.985
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr6_-_7910716
|
1.978
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr17_+_15848422
|
1.967
|
|
ADORA2B
|
adenosine A2b receptor
|
|
chr19_-_4066739
|
1.964
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr17_+_45000483
|
1.952
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr1_-_24126031
|
1.947
|
NM_001127621
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr15_-_55880508
|
1.898
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr16_-_30569505
|
1.898
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr3_-_57583080
|
1.884
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr11_+_118443140
|
1.884
|
|
ARCN1
|
archain 1
|
|
chr19_+_1026608
|
1.874
|
|
CNN2
|
calponin 2
|
|
chr2_-_15701414
|
1.872
|
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr9_+_35732629
|
1.870
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr14_-_39572278
|
1.867
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr3_-_57582866
|
1.861
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr19_+_45394569
|
1.854
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr9_-_35072643
|
1.854
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr22_+_38864015
|
1.848
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr15_-_42264687
|
1.844
|
|
EHD4
|
EH-domain containing 4
|
|
chr3_-_57583090
|
1.834
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr11_+_118443091
|
1.828
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr19_+_1026627
|
1.811
|
|
CNN2
|
calponin 2
|
|
chr16_-_28222700
|
1.811
|
|
XPO6
|
exportin 6
|
|
chr9_-_35072596
|
1.802
|
|
VCP
|
valosin containing protein
|
|
chr3_-_128369525
|
1.786
|
|
RPN1
|
ribophorin I
|
|
chr22_+_45148437
|
1.785
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr19_-_54619025
|
1.758
|
NM_013342
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr3_+_132379204
|
1.732
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr11_+_128563993
|
1.726
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr11_-_2018568
|
1.724
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr16_+_88636818
|
1.699
|
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr2_+_234263153
|
1.685
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chrX_+_68725077
|
1.676
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr15_+_79603497
|
1.671
|
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr6_-_90062289
|
1.666
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr7_-_54826899
|
1.664
|
NM_001012456
NM_014302
|
SEC61G
|
Sec61 gamma subunit
|
|
chr10_-_120840296
|
1.663
|
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A
|
|
chr9_-_101984233
|
1.659
|
NM_033087
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chr11_+_65292576
|
1.654
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr3_+_132379130
|
1.650
|
NM_024818
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr8_-_81083660
|
1.642
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr2_-_75062214
|
1.634
|
|
|
|
|
chr15_+_79603427
|
1.632
|
NM_007364
|
TMED3
|
transmembrane emp24 protein transport domain containing 3
|
|
chr10_-_16859336
|
1.616
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr17_+_14204326
|
1.611
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr2_-_242212244
|
1.599
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr5_-_89825292
|
1.596
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr6_+_7107986
|
1.573
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr8_+_6565870
|
1.552
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr20_+_25228698
|
1.547
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr12_-_133404950
|
1.546
|
|
GOLGA3
|
golgin A3
|
|
chr2_-_118771663
|
1.546
|
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr7_+_128095881
|
1.540
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chr11_+_47430045
|
1.537
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr5_+_34656367
|
1.512
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_121711980
|
1.509
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr7_-_148725764
|
1.483
|
NM_004911
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr5_+_172571444
|
1.482
|
NM_001205
NM_013978
NM_013979
NM_013980
|
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1
|
|
chr19_+_39421562
|
1.479
|
NM_021107
NM_033363
|
MRPS12
|
mitochondrial ribosomal protein S12
|
|
chr4_+_110736636
|
1.462
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr11_+_47430131
|
1.460
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr12_-_53473171
|
1.446
|
NM_032840
|
SPRYD3
|
SPRY domain containing 3
|
|
chr3_+_132378735
|
1.445
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr21_-_44299660
|
1.444
|
NM_018669
NM_033661
|
WDR4
|
WD repeat domain 4
|
|
chr2_+_234263141
|
1.444
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|