|
chrX_+_43515406
|
4.029
|
NM_000240
|
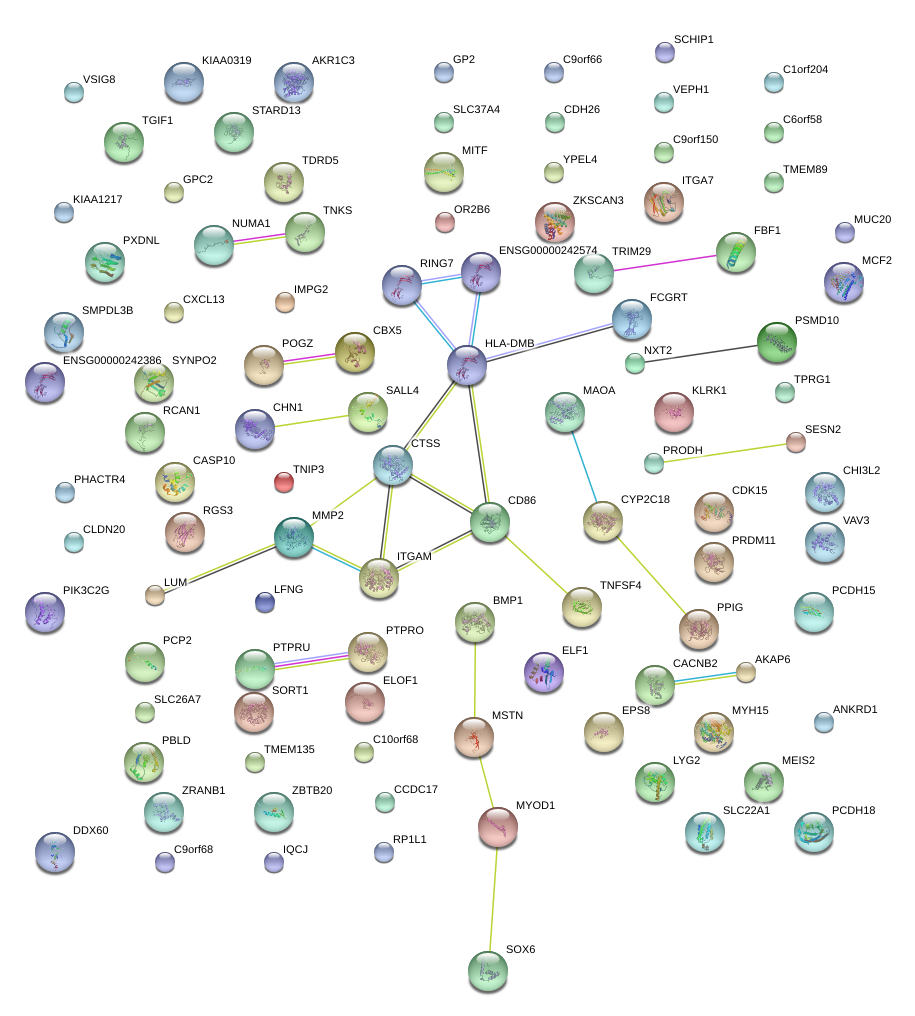
MAOA
|
monoamine oxidase A
|
|
chrX_+_43515441
|
3.430
|
|
MAOA
|
monoamine oxidase A
|
|
chr2_-_190927321
|
2.477
|
NM_005259
|
MSTN
|
myostatin
|
|
chr3_+_188664939
|
1.840
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chrX_+_43515531
|
1.712
|
|
MAOA
|
monoamine oxidase A
|
|
chr1_-_108231122
|
1.698
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr9_+_12774988
|
1.528
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr12_-_57674251
|
1.506
|
|
|
|
|
chr10_-_92681025
|
1.415
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr11_-_57417247
|
1.319
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr7_+_2557496
|
1.276
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr22_-_18923963
|
1.236
|
NM_001195226
NM_016335
|
PRODH
|
proline dehydrogenase (oxidase) 1
|
|
chr12_-_57693146
|
1.212
|
|
|
|
|
chr10_-_56560947
|
1.046
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr10_-_70066594
|
1.038
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr15_-_37393364
|
0.990
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr3_+_69985750
|
0.979
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_157213138
|
0.893
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr19_-_7698569
|
0.857
|
NM_174895
|
PCP2
|
Purkinje cell protein 2
|
|
chr8_+_92261515
|
0.805
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr2_-_175712276
|
0.780
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr3_-_101039403
|
0.687
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr3_+_121774208
|
0.657
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr4_-_122137781
|
0.628
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr3_-_48659188
|
0.624
|
NM_001008269
|
TMEM89
|
transmembrane protein 89
|
|
chr8_+_9413390
|
0.597
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr8_+_9413422
|
0.575
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr1_+_179560747
|
0.564
|
NM_001199091
NM_001199085
NM_173533
|
TDRD5
|
tudor domain containing 5
|
|
chr6_+_127898318
|
0.553
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr11_+_27015668
|
0.537
|
|
|
|
|
chr1_-_150738232
|
0.495
|
|
CTSS
|
cathepsin S
|
|
chr1_-_159832311
|
0.487
|
NM_001013661
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
|
chr19_+_45116921
|
0.477
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr11_-_16497917
|
0.472
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_+_87032428
|
0.462
|
|
TMEM135
|
transmembrane protein 135
|
|
chr13_-_34250899
|
0.454
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr21_-_35899046
|
0.447
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr6_+_155585146
|
0.421
|
NM_001001346
|
CLDN20
|
claudin 20
|
|
chr10_+_32856650
|
0.414
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr8_-_10512616
|
0.394
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr3_-_108248168
|
0.393
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr9_-_4666665
|
0.383
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr1_-_46089638
|
0.380
|
NM_001114938
NM_001190182
|
CCDC17
|
coiled-coil domain containing 17
|
|
chr3_+_195447751
|
0.371
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr17_-_73937105
|
0.354
|
NM_001080542
|
FBF1
|
Fas (TNFRSF6) binding factor 1
|
|
chr12_-_15815073
|
0.339
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr16_+_55515468
|
0.337
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr11_-_120008801
|
0.309
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr20_+_58571416
|
0.305
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chr1_+_111770280
|
0.291
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr12_-_91505215
|
0.288
|
|
LUM
|
lumican
|
|
chr10_+_18689512
|
0.283
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr1_+_28261503
|
0.265
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr1_+_28764660
|
0.251
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr18_+_3447607
|
0.239
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr10_+_5136613
|
0.227
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_24755440
|
0.226
|
|
KIAA1217
|
KIAA1217
|
|
chr12_+_18414473
|
0.219
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr11_-_71752099
|
0.217
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr2_+_202047620
|
0.215
|
NM_001206524
NM_001206542
NM_001230
NM_032974
NM_032976
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr1_-_173174611
|
0.202
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr11_-_118900078
|
0.195
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr9_+_66457288
|
0.191
|
|
|
|
|
chr6_-_32908583
|
0.189
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr11_+_45115563
|
0.186
|
NM_020229
|
PRDM11
|
PR domain containing 11
|
|
chr6_+_28317687
|
0.180
|
NM_001242894
NM_001242895
NM_024493
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr4_+_78432905
|
0.179
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr1_+_28585959
|
0.177
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr1_-_151414641
|
0.173
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr7_-_99774821
|
0.158
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr3_-_157221362
|
0.153
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr10_+_24755459
|
0.151
|
|
KIAA1217
|
KIAA1217
|
|
chr1_-_151431648
|
0.150
|
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr4_-_169239816
|
0.149
|
NM_017631
|
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60
|
|
chr6_+_160542862
|
0.145
|
NM_003057
NM_153187
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr11_-_120008738
|
0.143
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr10_+_18549583
|
0.142
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr14_+_32798478
|
0.141
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr9_+_116356199
|
0.137
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr10_+_96443250
|
0.134
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr2_-_99871569
|
0.134
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr16_-_20338808
|
0.130
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr4_+_119809978
|
0.126
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr11_+_17741109
|
0.123
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr8_-_52721903
|
0.123
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr6_-_24583454
|
0.120
|
NM_001252328
|
KIAA0319
|
KIAA0319
|
|
chr9_-_215892
|
0.119
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr7_+_37723378
|
0.118
|
|
|
|
|
chr20_-_50418946
|
0.111
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chrX_-_138790328
|
0.100
|
NM_001099855
NM_001171876
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr7_+_141811508
|
0.087
|
|
LOC93432
|
maltase-glucoamylase (alpha-glucosidase) pseudogene
|
|
chr2_+_202671197
|
0.087
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr3_-_114343791
|
0.086
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_+_15699277
|
0.084
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr6_+_27925018
|
0.078
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr7_+_102023016
|
0.078
|
|
|
|
|
chr4_-_138453628
|
0.073
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr3_+_159557649
|
0.071
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_56106061
|
0.068
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chrX_+_108779009
|
0.066
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr10_+_126630691
|
0.066
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr13_-_41593405
|
0.062
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_-_54653369
|
0.060
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr1_-_109935869
|
0.053
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chrX_-_107334749
|
0.050
|
NM_002814
NM_170750
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10
|
|
chr19_+_50016490
|
0.047
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr16_+_31271311
|
0.047
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr16_-_73092533
|
0.047
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr19_+_21324826
|
0.045
|
NM_133473
|
ZNF431
|
zinc finger protein 431
|
|
chr7_-_44580857
|
0.036
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr7_+_94292737
|
0.033
|
|
PEG10
|
paternally expressed 10
|
|
chr12_-_4488707
|
0.031
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr5_-_9630462
|
0.031
|
NM_019599
|
TAS2R1
|
taste receptor, type 2, member 1
|
|
chrX_-_64196268
|
0.029
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr12_-_52604488
|
0.028
|
NM_001242696
|
LOC283403
|
uncharacterized LOC283403
|
|
chr7_+_99905324
|
0.026
|
NM_001004351
|
SPDYE3
|
speedy homolog E3 (Xenopus laevis)
|
|
chr13_-_36050758
|
0.025
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr17_-_29624249
|
0.024
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr20_+_43029865
|
0.023
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr11_+_60048013
|
0.023
|
NM_001243266
NM_024021
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chr11_-_129062092
|
0.022
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_65159566
|
0.022
|
|
LOC400958
|
uncharacterized LOC400958
|
|
chrX_-_23925947
|
0.020
|
NM_024122
|
APOO
|
apolipoprotein O
|
|
chr1_+_222910557
|
0.017
|
NM_207468
|
FAM177B
|
family with sequence similarity 177, member B
|
|
chr1_-_27953046
|
0.017
|
NM_001042729
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr22_+_41957011
|
0.017
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr20_-_52199635
|
0.016
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr11_-_128737132
|
0.011
|
NM_153764
NM_153765
NM_153766
NM_153767
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr6_+_37403405
|
0.011
|
|
|
|
|
chr1_-_152386727
|
0.006
|
NM_016190
|
CRNN
|
cornulin
|
|
chr1_-_68697997
|
0.005
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_+_3668961
|
0.005
|
NM_152492
|
CCDC27
|
coiled-coil domain containing 27
|
|
chr12_-_57328044
|
0.003
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr2_+_29038862
|
0.002
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr1_-_173933176
|
0.002
|
|
RC3H1
|
ring finger and CCCH-type domains 1
|