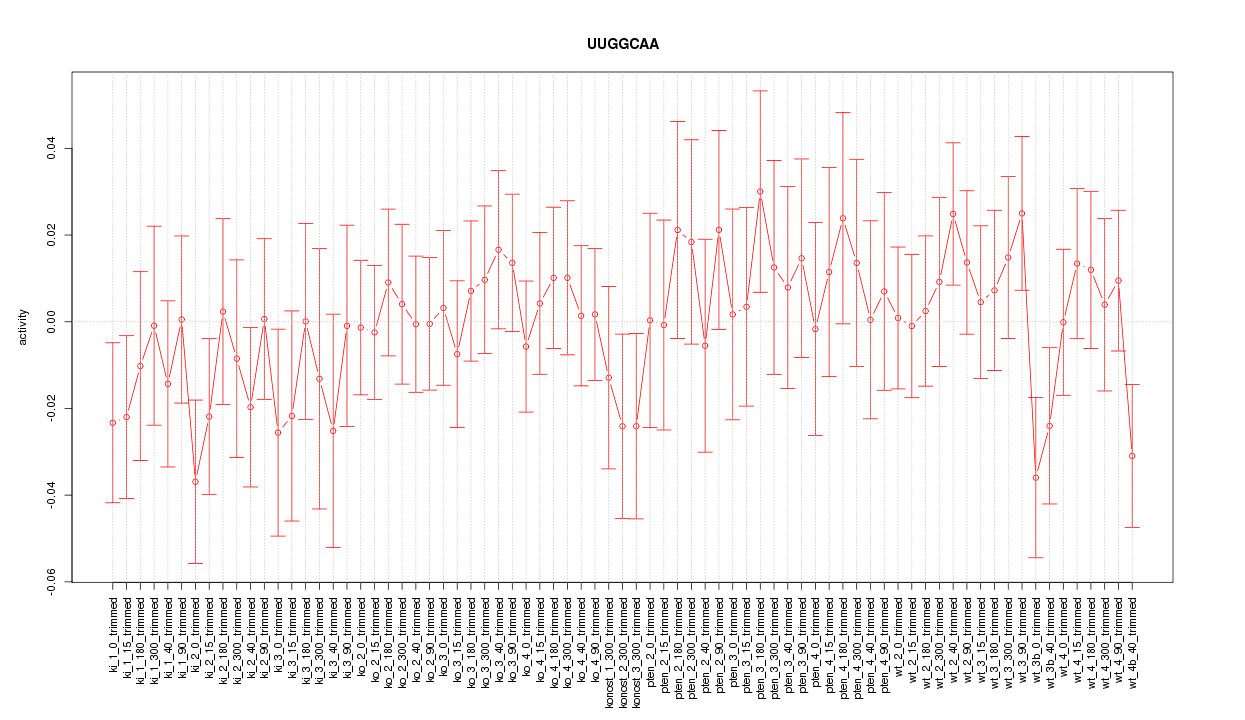
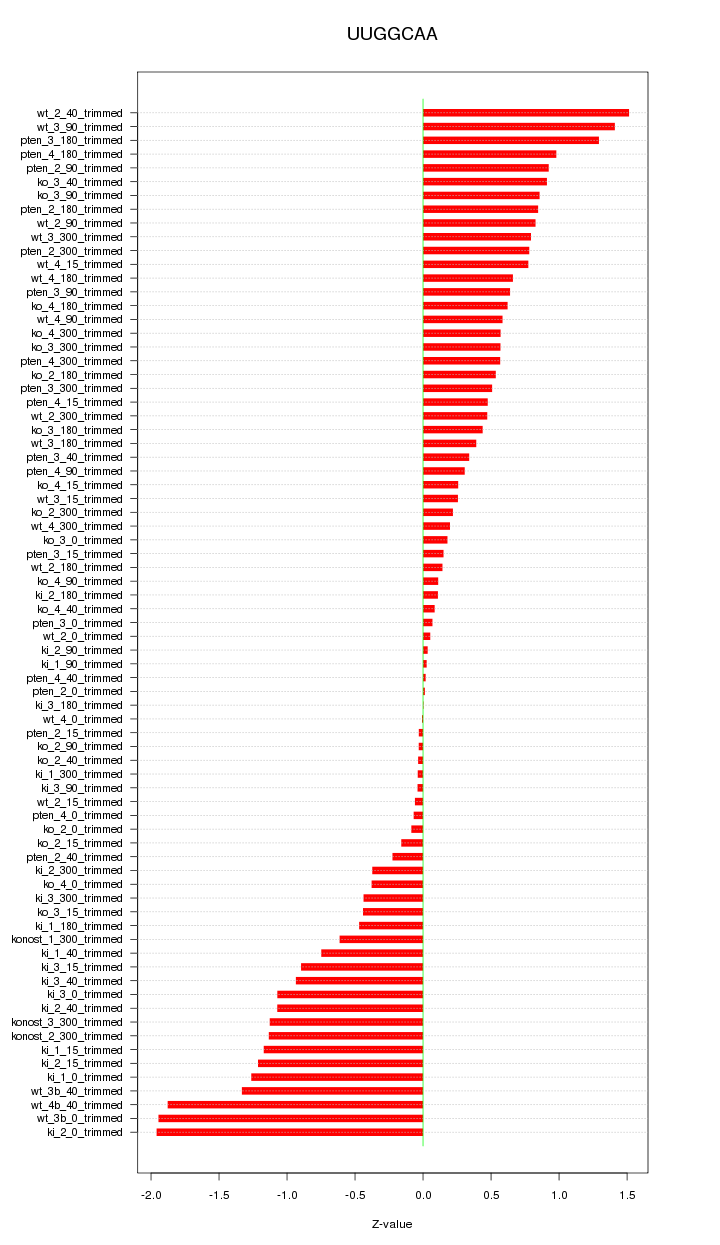
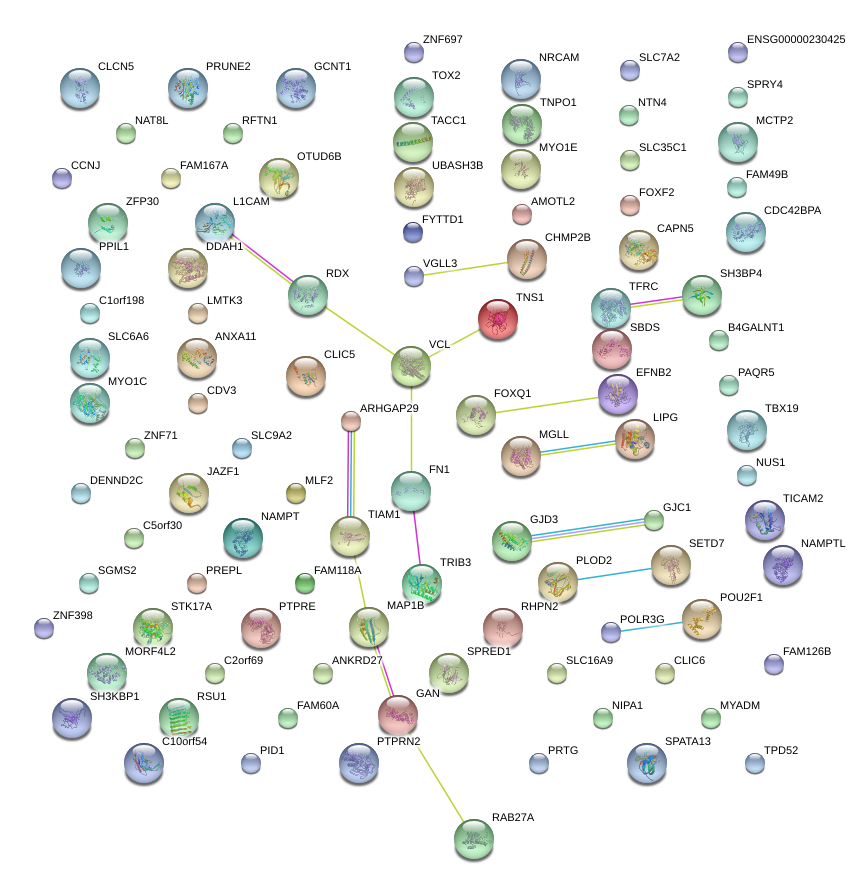
|
chr8_-_11324184
|
4.774
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr19_+_54369610
|
4.294
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54372659
|
4.100
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_54371125
|
3.931
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr8_+_17396285
|
3.880
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr2_+_103236147
|
3.603
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr8_+_17354562
|
3.465
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr3_-_87040246
|
3.373
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr9_+_79093256
|
3.131
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79115548
|
3.037
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79056581
|
3.013
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr5_-_141704575
|
2.904
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr2_-_230135954
|
2.852
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr9_+_79074067
|
2.700
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr4_+_108814419
|
2.605
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr4_+_108745718
|
2.484
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_-_86043932
|
2.437
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_+_97803064
|
2.389
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr7_-_107880492
|
2.367
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr1_-_94703252
|
2.321
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chrX_-_153141398
|
2.248
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr7_-_108096693
|
2.207
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr18_+_47088400
|
2.188
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr15_+_38544966
|
2.163
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr15_+_69606741
|
2.080
|
NM_001104554
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr11_+_45826640
|
2.021
|
NM_018389
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr11_+_45825622
|
1.997
|
NM_001145265
NM_001145266
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr20_+_361272
|
1.966
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr15_+_69591272
|
1.955
|
NM_017705
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr4_+_2061238
|
1.928
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr19_+_57106622
|
1.892
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chrX_+_49832214
|
1.873
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_-_85930733
|
1.818
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_-_158380360
|
1.804
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr6_-_45983279
|
1.801
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr10_+_75757847
|
1.794
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr7_-_105925410
|
1.731
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr1_-_120190389
|
1.723
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr3_-_195808997
|
1.716
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr15_-_56035176
|
1.702
|
NM_173814
|
PRTG
|
protogenin
|
|
chr8_+_92082423
|
1.643
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chr1_-_230991781
|
1.638
|
NM_001136495
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr6_+_11094265
|
1.628
|
NM_001135575
|
C6orf228
|
chromosome 6 open reading frame 228
|
|
chr21_-_32931289
|
1.607
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr3_-_195808958
|
1.604
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_-_231005334
|
1.594
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr10_+_129845806
|
1.578
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr3_+_197476623
|
1.531
|
NM_032288
|
FYTTD1
|
forty-two-three domain containing 1
|
|
chr10_-_61469322
|
1.520
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr9_-_79520878
|
1.483
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr2_-_44587812
|
1.473
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr7_+_43622638
|
1.469
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr2_-_201936389
|
1.462
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr19_-_49016418
|
1.455
|
NM_001080434
|
LMTK3
|
lemur tyrosine kinase 3
|
|
chr7_-_66460403
|
1.453
|
NM_016038
|
SBDS
|
Shwachman-Bodian-Diamond syndrome
|
|
chr2_-_218808705
|
1.450
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr5_-_114938060
|
1.444
|
NM_021649
|
TICAM2
|
toll-like receptor adaptor molecule 2
|
|
chr5_+_89770680
|
1.438
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr13_+_24734788
|
1.432
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr7_+_148823475
|
1.426
|
NM_020781
|
ZNF398
|
zinc finger protein 398
|
|
chr3_-_127541160
|
1.421
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr5_+_72143929
|
1.409
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr3_+_197476423
|
1.408
|
NM_001011537
|
FYTTD1
|
forty-two-three domain containing 1
|
|
chr4_-_140477290
|
1.400
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr17_-_42908178
|
1.399
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr12_-_58026846
|
1.398
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr3_+_133292404
|
1.366
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_-_216300770
|
1.364
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr3_-_145879222
|
1.359
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr12_-_31479120
|
1.354
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr22_+_45704821
|
1.348
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr1_+_200708604
|
1.347
|
NM_203459
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2
|
|
chr11_-_110167191
|
1.335
|
NM_002906
|
RDX
|
radixin
|
|
chr15_-_55562483
|
1.333
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr3_-_127541975
|
1.308
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr3_+_133293256
|
1.308
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_+_200775974
|
1.307
|
NM_153689
|
C2orf69
|
chromosome 2 open reading frame 69
|
|
chr1_-_115212628
|
1.294
|
NM_198459
|
DENND2C
|
DENN/MADD domain containing 2C
|
|
chrX_-_102942968
|
1.291
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr8_-_130951996
|
1.287
|
NM_016623
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr8_-_80993009
|
1.278
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr17_-_1395950
|
1.272
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr6_+_1312674
|
1.263
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr3_-_16555128
|
1.256
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr3_+_14444083
|
1.255
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr10_-_16859408
|
1.233
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr6_+_117996616
|
1.227
|
NM_138459
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae)
|
|
chr19_-_38146288
|
1.222
|
NM_014898
|
ZFP30
|
zinc finger protein 30 homolog (mouse)
|
|
chrX_+_49687212
|
1.213
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr10_+_129705299
|
1.211
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr7_+_148844440
|
1.209
|
NM_170686
|
ZNF398
|
zinc finger protein 398
|
|
chr5_+_71403040
|
1.184
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr12_-_96184510
|
1.183
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr20_+_42574535
|
1.175
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr10_-_73533196
|
1.169
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_+_167298280
|
1.168
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_-_33555757
|
1.166
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr10_-_81965327
|
1.166
|
NM_001157
NM_145868
NM_145869
|
ANXA11
|
annexin A11
|
|
chr2_+_235860616
|
1.163
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr11_+_76777926
|
1.159
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr16_+_81348564
|
1.143
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr15_+_94841429
|
1.142
|
NM_001159643
NM_018349
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr7_-_28220342
|
1.136
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr15_-_23086842
|
1.136
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr1_-_231004173
|
1.134
|
NM_032800
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chrX_-_19688991
|
1.126
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr13_-_107187312
|
1.121
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chrX_-_19817907
|
1.117
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_227505796
|
1.116
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr6_+_1390068
|
1.116
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr11_+_122526339
|
1.111
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr8_+_38585703
|
1.109
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr5_+_102594406
|
1.106
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr6_-_36842799
|
1.101
|
NM_016059
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1
|
|
chr3_+_87276398
|
1.099
|
NM_001244644
NM_014043
|
CHMP2B
|
charged multivesicular body protein 2B
|
|
chr3_-_134093235
|
1.096
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr15_-_55563091
|
1.084
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr19_-_33166057
|
1.083
|
NM_032139
|
ANKRD27
|
ankyrin repeat domain 27 (VPS9 domain)
|
|
chr20_+_42544781
|
1.082
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr3_-_133614421
|
1.074
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr3_+_32433151
|
1.073
|
NM_138410
NM_181472
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr6_+_80340971
|
1.070
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr15_-_23086290
|
1.069
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr6_+_64281910
|
1.061
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_-_44586854
|
1.061
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr11_-_47516072
|
1.059
|
NM_198700
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr6_-_57086204
|
1.057
|
NM_183227
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr4_+_71768056
|
1.050
|
NM_001244766
NM_001244767
NM_173468
|
MOB1B
|
MOB kinase activator 1B
|
|
chr17_-_42907606
|
1.046
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr19_+_34663311
|
1.039
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr15_+_63481713
|
1.036
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr5_+_154092461
|
1.030
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr2_-_44588622
|
1.029
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr3_-_27763784
|
1.027
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr19_-_10341947
|
1.026
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr5_-_59783885
|
1.024
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_+_138483024
|
1.023
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr2_-_44588943
|
1.021
|
NM_001171603
NM_001171617
|
PREPL
|
prolyl endopeptidase-like
|
|
chr9_+_119449573
|
1.019
|
NM_001099679
NM_012210
|
TRIM32
|
tripartite motif containing 32
|
|
chr22_-_22307212
|
1.015
|
NM_014634
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr1_-_153940187
|
1.008
|
NM_014437
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr2_+_109204904
|
1.007
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr11_+_111895537
|
1.000
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr6_-_57087018
|
0.998
|
NM_016277
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr9_-_131940538
|
0.998
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr15_-_55581969
|
0.998
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr4_-_184580104
|
0.995
|
NM_152682
|
RWDD4
|
RWD domain containing 4
|
|
chr6_-_91006460
|
0.992
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr8_+_123793894
|
0.983
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr16_+_85061303
|
0.980
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr17_+_37617688
|
0.979
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr19_-_48894755
|
0.977
|
NM_006801
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr1_+_90098643
|
0.976
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_-_23495347
|
0.971
|
NM_001142546
NM_033631
|
LUZP1
|
leucine zipper protein 1
|
|
chr7_+_94539209
|
0.966
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr5_+_138678130
|
0.961
|
NM_001033112
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr11_+_107879407
|
0.960
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr2_+_208394615
|
0.954
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr15_+_44829265
|
0.953
|
NM_003758
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr3_+_187943192
|
0.953
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_111717585
|
0.946
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr2_-_101034049
|
0.943
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr15_-_49255640
|
0.925
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr5_+_86564727
|
0.923
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr8_-_57906423
|
0.922
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr7_-_75988315
|
0.921
|
NM_012479
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr4_+_41614911
|
0.914
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_+_45560529
|
0.914
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr17_-_27621163
|
0.913
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr13_-_50367027
|
0.910
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr2_+_48541767
|
0.909
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr12_-_104234968
|
0.897
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr3_+_57261729
|
0.895
|
NM_012096
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr5_-_43557520
|
0.890
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chr20_+_42543417
|
0.881
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr16_+_71879893
|
0.878
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr14_+_55221505
|
0.876
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr10_+_95653668
|
0.875
|
NM_001134658
NM_153226
|
SLC35G1
|
solute carrier family 35, member G1
|
|
chr5_-_95297597
|
0.874
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr4_+_39699663
|
0.871
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr14_+_103801160
|
0.866
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_+_178694273
|
0.857
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_-_95392501
|
0.848
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr6_-_91006580
|
0.843
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr5_+_72112417
|
0.830
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr14_+_103800492
|
0.828
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr14_+_105331581
|
0.827
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr7_-_127670995
|
0.826
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr5_-_126366439
|
0.823
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr5_+_31799030
|
0.819
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr6_-_16761600
|
0.819
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr1_+_198125890
|
0.816
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr7_+_18535351
|
0.816
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr11_-_47510518
|
0.815
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr3_-_27498244
|
0.811
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr6_-_30710219
|
0.809
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr13_-_20735182
|
0.806
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr3_+_111718006
|
0.805
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr6_-_119031230
|
0.804
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr22_+_45705766
|
0.803
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|