|
chr19_+_45971249
|
12.962
|
NM_001114171
NM_006732
|
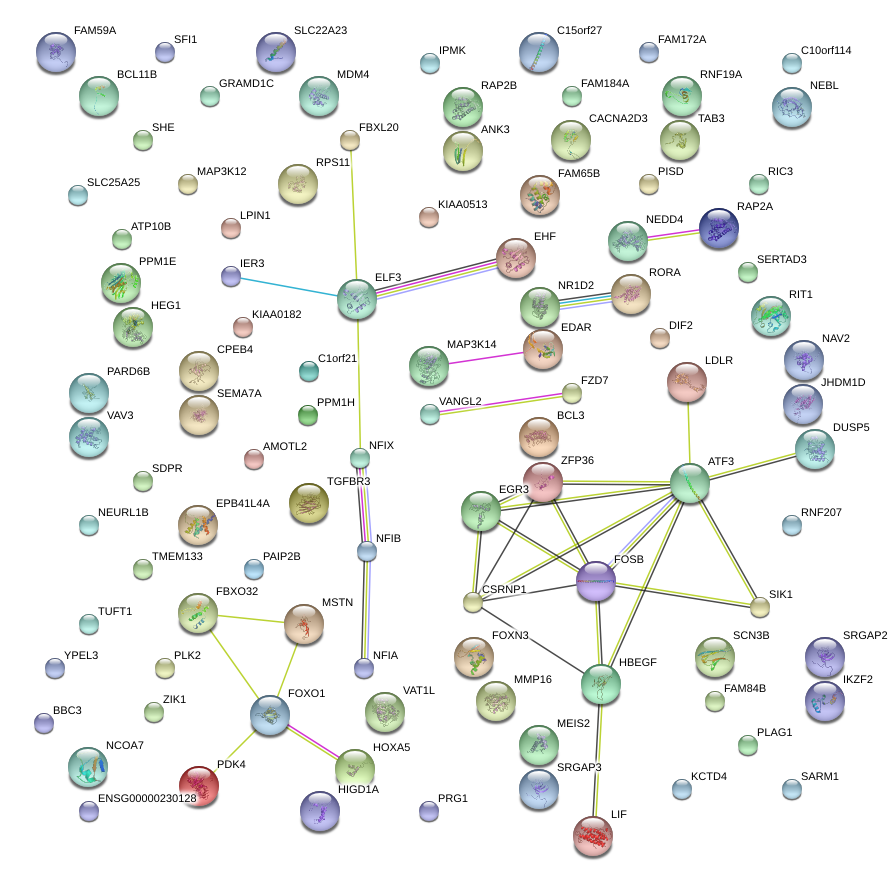
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr6_+_126240369
|
10.069
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_+_212788359
|
9.090
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr6_+_126111782
|
9.025
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_+_212781969
|
8.630
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr6_+_126102278
|
7.524
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_+_212738675
|
7.402
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr8_-_22549901
|
6.182
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
6.134
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
6.071
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr10_+_112257624
|
5.445
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr8_-_124544376
|
5.271
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr5_-_57755787
|
5.154
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr8_-_124553448
|
4.515
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr5_-_139726173
|
4.391
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr11_+_19734821
|
4.385
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr11_+_34654010
|
4.166
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr3_-_39195101
|
4.020
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr11_+_34642587
|
3.914
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr11_+_19372270
|
3.911
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr19_+_45251933
|
3.899
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr9_+_130854127
|
3.663
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr2_+_11886739
|
3.429
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr9_+_130860816
|
3.406
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr1_-_92351613
|
3.370
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr9_+_130830447
|
3.289
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr16_+_77822400
|
3.251
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chr13_-_41240607
|
3.238
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr7_-_95225802
|
3.221
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr3_+_54156691
|
3.188
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr3_+_152880024
|
3.139
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr1_-_108507474
|
3.138
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_+_61547625
|
3.054
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_201979689
|
3.032
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr19_+_39897457
|
2.962
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr1_-_92371558
|
2.892
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_108231122
|
2.823
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr21_-_44846927
|
2.676
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr1_+_160370363
|
2.673
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr12_-_63328663
|
2.650
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr3_-_9291251
|
2.628
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr2_-_109605668
|
2.626
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr1_+_6266140
|
2.603
|
NM_207396
|
RNF207
|
ring finger protein 207
|
|
chr1_+_61542928
|
2.456
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr6_-_24911194
|
2.418
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr3_+_23986735
|
2.397
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_23987514
|
2.367
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr20_+_49348050
|
2.214
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr15_-_74726258
|
2.210
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr14_-_90085325
|
2.191
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr11_-_8190535
|
2.021
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr17_-_43394326
|
2.019
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr3_+_113616317
|
2.008
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr1_+_61547388
|
1.984
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_127570710
|
1.967
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr5_-_111754738
|
1.949
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr1_-_154474524
|
1.945
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr10_-_21186530
|
1.914
|
NM_006393
|
NEBL
|
nebulette
|
|
chr15_-_74726000
|
1.913
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr2_-_192711923
|
1.895
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr3_-_124774735
|
1.867
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr10_-_21463019
|
1.863
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr17_+_26698986
|
1.819
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr16_+_85646923
|
1.811
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr13_-_45775174
|
1.810
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr17_-_37557851
|
1.810
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr14_-_89883306
|
1.805
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr5_-_160279045
|
1.776
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr3_-_134093235
|
1.717
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr8_-_89339716
|
1.683
|
NM_005941
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chr3_+_113557679
|
1.670
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr10_-_21786205
|
1.635
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr16_+_85645014
|
1.631
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr2_-_71454200
|
1.607
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr8_-_57123858
|
1.601
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr6_-_119399759
|
1.584
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr1_+_184356128
|
1.579
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr16_+_85061303
|
1.573
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr7_-_27183225
|
1.559
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr6_-_30712294
|
1.553
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr11_-_123525300
|
1.550
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr2_-_190927321
|
1.529
|
NM_005259
|
MSTN
|
myostatin
|
|
chr10_-_62493282
|
1.518
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_11200037
|
1.516
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr17_+_56833231
|
1.508
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr15_-_56285732
|
1.506
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr7_-_139876718
|
1.498
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr10_-_61900659
|
1.493
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_3456734
|
1.486
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr2_-_214016332
|
1.452
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr19_-_40948504
|
1.409
|
NM_013368
|
SERTAD3
|
SERTA domain containing 3
|
|
chr5_-_93447237
|
1.407
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr15_-_37390372
|
1.398
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr22_-_32026783
|
1.397
|
NM_014338
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr9_-_14314036
|
1.388
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr19_-_47735858
|
1.379
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr10_-_62332666
|
1.374
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_60027630
|
1.369
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr2_+_202899309
|
1.365
|
NM_003507
|
FZD7
|
frizzled family receptor 7
|
|
chr1_+_151512760
|
1.365
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|
|
chr5_+_172068188
|
1.339
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr15_+_76352270
|
1.339
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr12_-_53893195
|
1.333
|
NM_001193511
NM_006301
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chr9_-_14398981
|
1.324
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr15_-_60884615
|
1.314
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_+_173315313
|
1.305
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chrX_-_30907408
|
1.296
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr1_-_155881163
|
1.293
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr18_-_30050394
|
1.288
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr6_-_119470336
|
1.285
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr22_-_30642683
|
1.280
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_+_204485461
|
1.264
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr15_-_37391596
|
1.252
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr4_-_16228097
|
1.246
|
NM_153365
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr1_-_47655723
|
1.238
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr3_-_155394104
|
1.237
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr17_+_43971655
|
1.219
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr10_-_62149487
|
1.216
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_+_58939539
|
1.204
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr17_+_25621105
|
1.192
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr14_+_56584854
|
1.191
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr3_+_150804675
|
1.188
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr22_+_31608249
|
1.183
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr19_-_40950238
|
1.170
|
NM_203344
|
SERTAD3
|
SERTA domain containing 3
|
|
chr10_+_18429605
|
1.164
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr3_-_155421996
|
1.152
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr19_+_57154526
|
1.147
|
NM_001193628
|
LOC147670
|
uncharacterized LOC147670
|
|
chr3_+_69915374
|
1.146
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_41221530
|
1.139
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr12_-_123451031
|
1.134
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr1_+_211433285
|
1.128
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr11_+_34937676
|
1.122
|
NM_001135024
NM_001166158
NM_003477
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr6_-_3445198
|
1.119
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr1_+_213123853
|
1.113
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr3_-_178789597
|
1.094
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chrX_+_9754465
|
1.083
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr20_-_30795346
|
1.053
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chrX_+_56258785
|
1.047
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr9_+_137218313
|
1.044
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr12_-_54785018
|
1.040
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr7_-_139477437
|
1.040
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr19_-_47734215
|
1.025
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr3_-_105587710
|
1.017
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr19_+_38397841
|
1.014
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr3_+_69788562
|
1.009
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr14_+_70078309
|
1.007
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr15_-_51914770
|
1.006
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr16_-_70719854
|
1.003
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr15_-_37392358
|
1.002
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr19_-_36391448
|
0.999
|
NM_139239
|
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chr8_+_125486949
|
0.996
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr14_+_33408458
|
0.994
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr2_+_7057471
|
0.990
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr10_+_1102741
|
0.973
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr1_+_40420781
|
0.972
|
NM_001136493
NM_032793
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr3_-_15563160
|
0.970
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr6_-_42419782
|
0.969
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr6_-_33160199
|
0.969
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr6_+_142622797
|
0.964
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr10_+_63661012
|
0.964
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr17_-_71088832
|
0.956
|
NM_001159770
NM_139177
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr1_+_13910251
|
0.952
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr19_-_46234126
|
0.944
|
NM_001080469
|
FBXO46
|
F-box protein 46
|
|
chr3_+_113465860
|
0.943
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr7_+_145813452
|
0.935
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr20_-_20693122
|
0.919
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr2_+_169312793
|
0.918
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr2_+_110371887
|
0.915
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr12_-_92539614
|
0.913
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr3_+_69812620
|
0.913
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_43732358
|
0.899
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr10_-_94050799
|
0.897
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr15_-_102192337
|
0.893
|
NM_025141
NM_078474
|
TM2D3
|
TM2 domain containing 3
|
|
chr12_-_31743822
|
0.889
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr1_-_161039745
|
0.887
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr15_-_37393364
|
0.887
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr3_+_69812961
|
0.869
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_-_38556682
|
0.864
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr16_-_23160509
|
0.864
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr2_-_214015053
|
0.854
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr17_-_2304217
|
0.852
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr2_+_219187858
|
0.844
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr21_+_18885104
|
0.838
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr16_-_25268841
|
0.832
|
NM_001012981
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2
|
|
chr1_-_179112178
|
0.829
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr8_+_17354562
|
0.827
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr18_-_74207145
|
0.825
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr10_+_35625608
|
0.816
|
NM_145012
|
CCNY
|
cyclin Y
|
|
chr6_-_143266283
|
0.803
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr16_-_1429612
|
0.802
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr17_+_47572592
|
0.800
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr1_-_179198814
|
0.788
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr5_-_38595506
|
0.782
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr8_+_38758752
|
0.776
|
NM_021623
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr8_-_62627167
|
0.775
|
NM_001164754
NM_001164755
NM_001164756
NM_004318
NM_032466
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr16_-_73082169
|
0.774
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr3_+_69985750
|
0.768
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr17_+_72983723
|
0.765
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr1_-_36023005
|
0.761
|
NM_024874
|
KIAA0319L
|
KIAA0319-like
|
|
chr6_-_119031230
|
0.756
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|