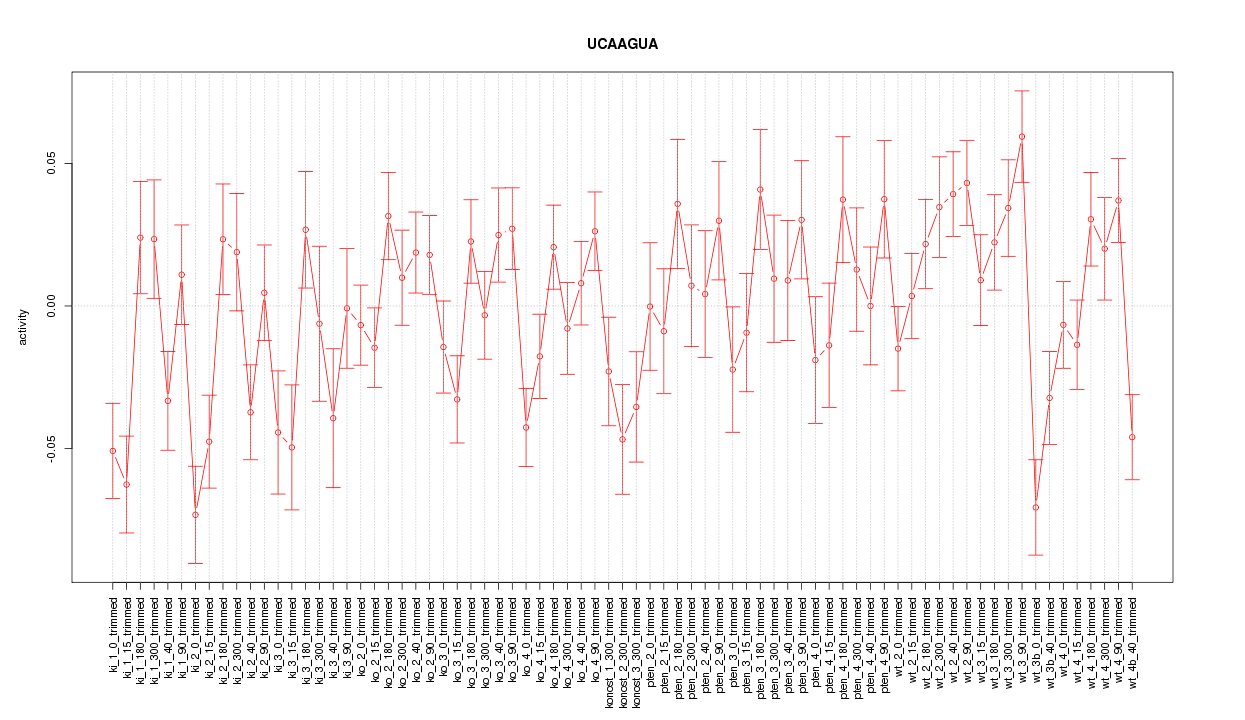
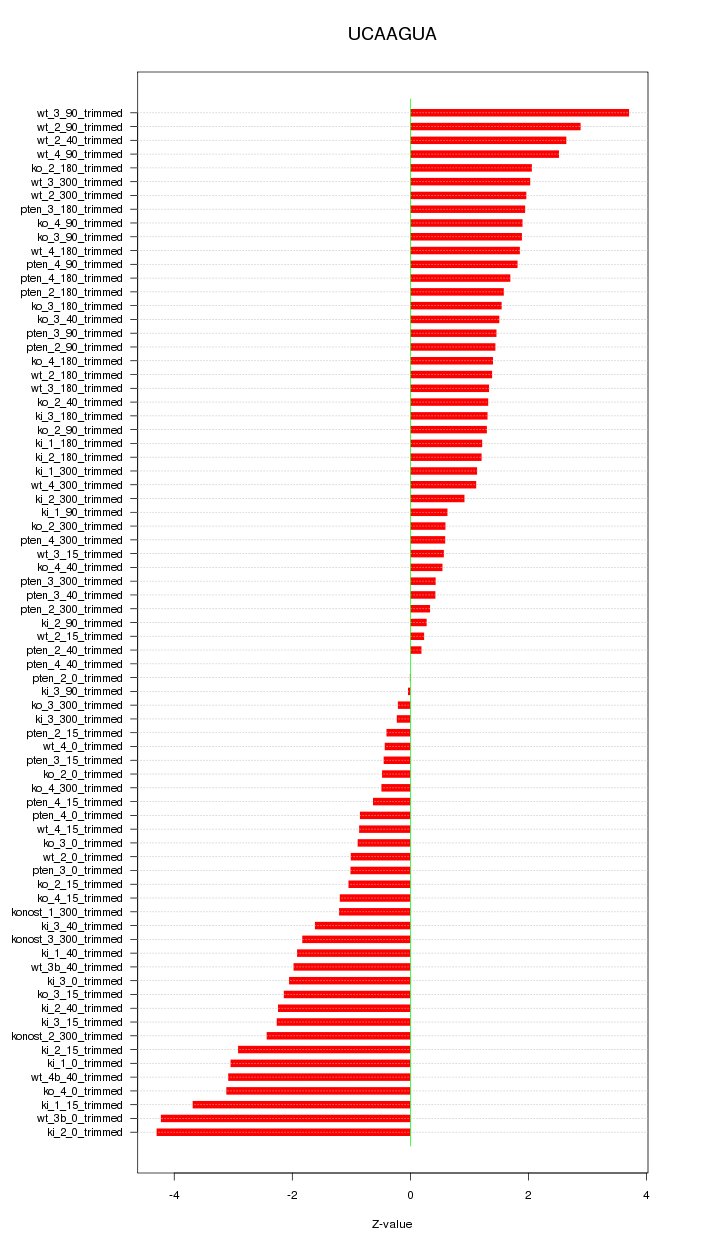
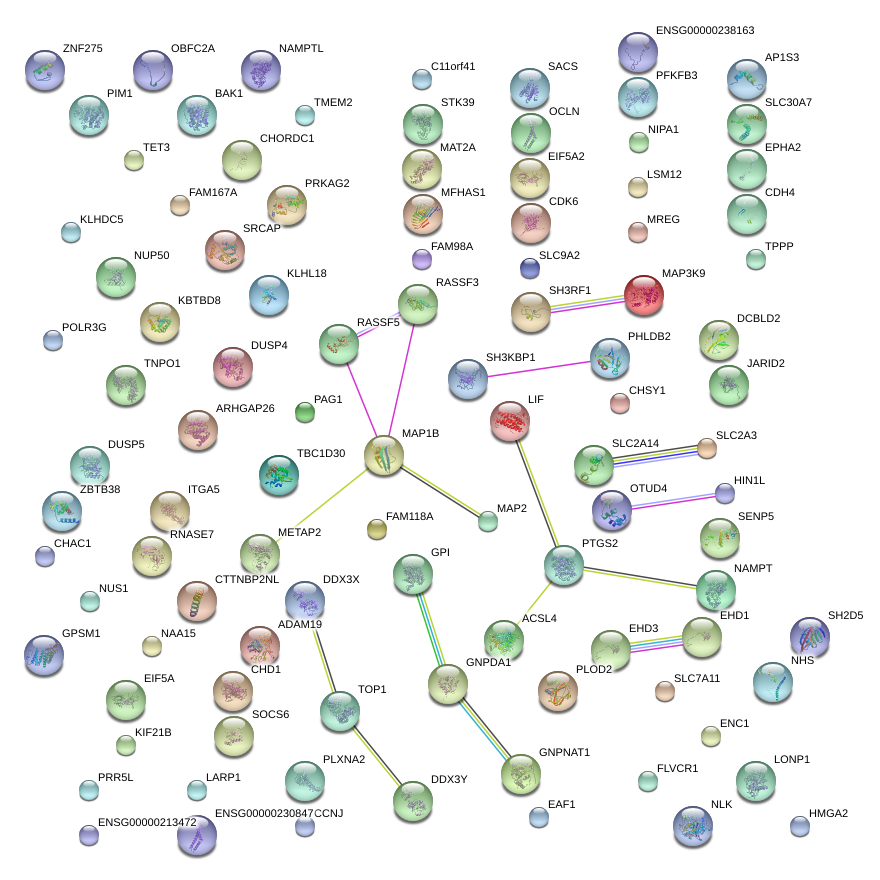
|
chr3_+_111578528
|
10.955
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr3_+_111578026
|
10.884
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr3_+_111451326
|
10.859
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr5_+_68800003
|
9.552
|
NM_001205255
|
OCLN
|
occludin
|
|
chr5_+_68788387
|
8.572
|
NM_001205254
|
OCLN
|
occludin
|
|
chr5_+_68788118
|
8.274
|
NM_002538
|
OCLN
|
occludin
|
|
chr10_+_97803064
|
8.012
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr9_-_74383301
|
7.722
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr2_+_103236147
|
7.375
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr10_+_6244839
|
6.513
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr4_-_139163222
|
6.338
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr1_-_186649421
|
6.271
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr10_+_6186758
|
6.224
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr12_+_65004191
|
5.997
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr2_-_216878314
|
5.976
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr8_-_29207795
|
5.652
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_+_71403040
|
5.648
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_693413
|
5.466
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr8_-_29206321
|
5.442
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr12_+_27933162
|
5.435
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr1_-_200992824
|
5.261
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr22_-_30642683
|
5.211
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr12_-_54813010
|
5.133
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr5_-_157002739
|
5.105
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr5_+_89770680
|
5.008
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr2_+_85766100
|
4.948
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr1_-_16482426
|
4.937
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr8_-_82024277
|
4.885
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr4_+_140222668
|
4.876
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr6_+_37137882
|
4.829
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr14_-_71275732
|
4.742
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr3_-_170626425
|
4.726
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr7_-_151511925
|
4.654
|
NM_001040633
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr3_+_67048726
|
4.653
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr2_+_74273449
|
4.644
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr6_-_33548017
|
4.556
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr11_+_36317724
|
4.536
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr8_-_11324184
|
4.505
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr17_+_26369617
|
4.500
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr9_+_139247508
|
4.488
|
NM_001145639
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr11_-_64646053
|
4.484
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr9_+_139249251
|
4.482
|
NM_001200003
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr15_+_41245635
|
4.465
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr5_-_73937248
|
4.413
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_+_36397534
|
4.392
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr1_-_21059132
|
4.289
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr7_-_151329153
|
4.199
|
NM_024429
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr10_+_112257624
|
4.186
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr3_+_196594708
|
4.081
|
NM_152699
|
SENP5
|
SUMO1/sentrin specific peptidase 5
|
|
chr18_+_67956136
|
4.067
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr3_+_15469052
|
3.962
|
NM_033083
|
EAF1
|
ELL associated factor 1
|
|
chr1_+_15272414
|
3.921
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr20_+_59827452
|
3.903
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr12_+_65218351
|
3.889
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr7_-_105925410
|
3.883
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr3_-_145879222
|
3.860
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr1_+_213031596
|
3.835
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chrX_-_108976509
|
3.822
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr3_+_141043054
|
3.803
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr12_+_66218141
|
3.759
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr2_+_192542860
|
3.716
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr7_-_92463211
|
3.662
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr11_-_89956531
|
3.576
|
NM_001144073
NM_012124
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1
|
|
chr11_+_36422546
|
3.530
|
NM_001160169
|
PRR5L
|
proline rich 5 like
|
|
chr13_-_24007822
|
3.511
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr5_+_154092461
|
3.485
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr5_+_72143929
|
3.434
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr5_-_98262217
|
3.423
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chrX_+_17653412
|
3.416
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr15_-_101792125
|
3.408
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr15_-_23086842
|
3.356
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr4_-_170192193
|
3.309
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr6_+_117996616
|
3.303
|
NM_138459
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae)
|
|
chrX_+_17393537
|
3.238
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr17_-_42144986
|
3.207
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr2_-_33824294
|
3.178
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr1_-_208417635
|
3.158
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr1_+_15256295
|
3.151
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr12_-_8088629
|
3.148
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr7_-_151574209
|
3.144
|
NM_016203
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr3_-_98620452
|
3.144
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chrX_-_19688991
|
3.117
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_101361589
|
3.105
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chrX_-_19817907
|
3.085
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_200708604
|
3.063
|
NM_203459
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2
|
|
chrX_+_152599544
|
3.048
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr4_+_166248816
|
3.032
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr5_+_142150291
|
3.000
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr14_-_53258329
|
2.975
|
NM_198066
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr15_-_23086290
|
2.966
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr2_-_169103886
|
2.955
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr4_-_146100793
|
2.930
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr6_+_15246299
|
2.925
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_-_224702300
|
2.886
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr11_+_33563772
|
2.877
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr22_+_45560529
|
2.876
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr1_+_112938799
|
2.875
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr22_+_45704821
|
2.874
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr8_-_8750739
|
2.836
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr3_+_47324329
|
2.822
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr20_+_39657461
|
2.757
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chrX_+_41192650
|
2.744
|
NM_001193416
NM_001193417
NM_001356
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked
|
|
chr14_+_21510384
|
2.722
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr2_+_210444364
|
2.715
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chrX_-_63425484
|
2.692
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr7_-_92465929
|
2.665
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr3_+_196466724
|
2.660
|
NM_002577
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr8_+_86157693
|
2.637
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr18_-_48724031
|
2.627
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr11_-_122932844
|
2.624
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr1_+_244216506
|
2.619
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_+_182756468
|
2.612
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr22_+_25960738
|
2.608
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr8_+_6565870
|
2.606
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr17_+_30813928
|
2.598
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr10_-_931694
|
2.579
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr3_-_119812973
|
2.572
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr8_-_130951996
|
2.564
|
NM_016623
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr22_+_45559725
|
2.542
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr8_-_23540401
|
2.531
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr2_+_208394615
|
2.511
|
NM_004379
NM_134442
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr14_+_67827030
|
2.473
|
NM_004094
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr16_+_69139466
|
2.466
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr4_-_71705188
|
2.452
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr2_+_210288668
|
2.451
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_-_161350304
|
2.447
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_-_64777703
|
2.443
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr16_+_69141442
|
2.425
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_-_179112178
|
2.420
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr4_+_120133764
|
2.420
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr1_-_51887740
|
2.403
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr5_-_78809658
|
2.396
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr15_-_49255640
|
2.387
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr12_+_70172748
|
2.375
|
NM_001024647
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr1_-_47080793
|
2.374
|
NM_145279
|
MOB3C
|
MOB kinase activator 3C
|
|
chr9_-_35115844
|
2.342
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr6_+_20402040
|
2.326
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr5_+_157170702
|
2.314
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr12_+_70133179
|
2.307
|
NM_175623
NM_175625
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr12_+_123868608
|
2.307
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr1_+_27153130
|
2.305
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr2_-_70529142
|
2.288
|
NM_032822
|
FAM136A
|
family with sequence similarity 136, member A
|
|
chr4_-_175443791
|
2.285
|
NM_000860
NM_001145816
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr1_-_94703252
|
2.283
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_+_5021096
|
2.272
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr5_-_59783885
|
2.271
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_120190389
|
2.271
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr4_-_109089444
|
2.268
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr6_-_167275657
|
2.258
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_+_18948276
|
2.252
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr12_-_46663207
|
2.233
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_-_67896098
|
2.224
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr9_+_139221931
|
2.220
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr20_+_36531498
|
2.220
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr6_+_17282931
|
2.165
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr5_+_72112417
|
2.162
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr12_-_77459198
|
2.159
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr1_+_244214552
|
2.156
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr15_-_25650647
|
2.147
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr3_-_33700932
|
2.147
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr6_+_144471631
|
2.134
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr4_+_41937105
|
2.127
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr11_-_10830581
|
2.120
|
NM_001042559
NM_001418
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr1_-_179198814
|
2.106
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr10_+_127408083
|
2.106
|
NM_001202438
NM_015608
|
C10orf137
|
chromosome 10 open reading frame 137
|
|
chrX_-_38186782
|
2.097
|
NM_000328
NM_001034853
|
RPGR
|
retinitis pigmentosa GTPase regulator
|
|
chr10_-_88281515
|
2.096
|
NM_015045
|
WAPAL
|
wings apart-like homolog (Drosophila)
|
|
chr1_-_115259433
|
2.092
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr1_+_15250578
|
2.083
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr5_-_58882274
|
2.075
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_5422792
|
2.070
|
NM_015325
|
KIAA0947
|
KIAA0947
|
|
chr1_-_201465700
|
2.067
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_201475966
|
2.067
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chrX_-_19905665
|
2.048
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_-_19748602
|
2.043
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr11_+_126081618
|
2.020
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr8_-_139926235
|
2.011
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr5_+_149340287
|
2.008
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr6_-_17706617
|
2.008
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr22_+_45705766
|
2.002
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr12_+_70132562
|
2.002
|
NM_022456
NM_175624
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr3_+_142838596
|
1.987
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr3_+_112051914
|
1.987
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr20_-_10654429
|
1.985
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr11_-_17191287
|
1.979
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr1_-_28415071
|
1.971
|
NM_001990
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chrX_-_138914384
|
1.970
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr15_-_52861177
|
1.958
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr20_+_43595119
|
1.950
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr1_+_100435539
|
1.928
|
NM_012243
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3
|
|
chr10_+_73723992
|
1.923
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr6_-_136788012
|
1.922
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_-_201476248
|
1.921
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr18_-_9614555
|
1.912
|
NM_001042388
NM_005134
|
PPP4R1
|
protein phosphatase 4, regulatory subunit 1
|
|
chr6_-_136847743
|
1.896
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr19_+_33864574
|
1.884
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr12_-_123834984
|
1.865
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr3_+_14444083
|
1.860
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr8_-_89339716
|
1.845
|
NM_005941
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chr5_-_58334936
|
1.841
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|