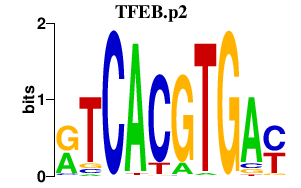
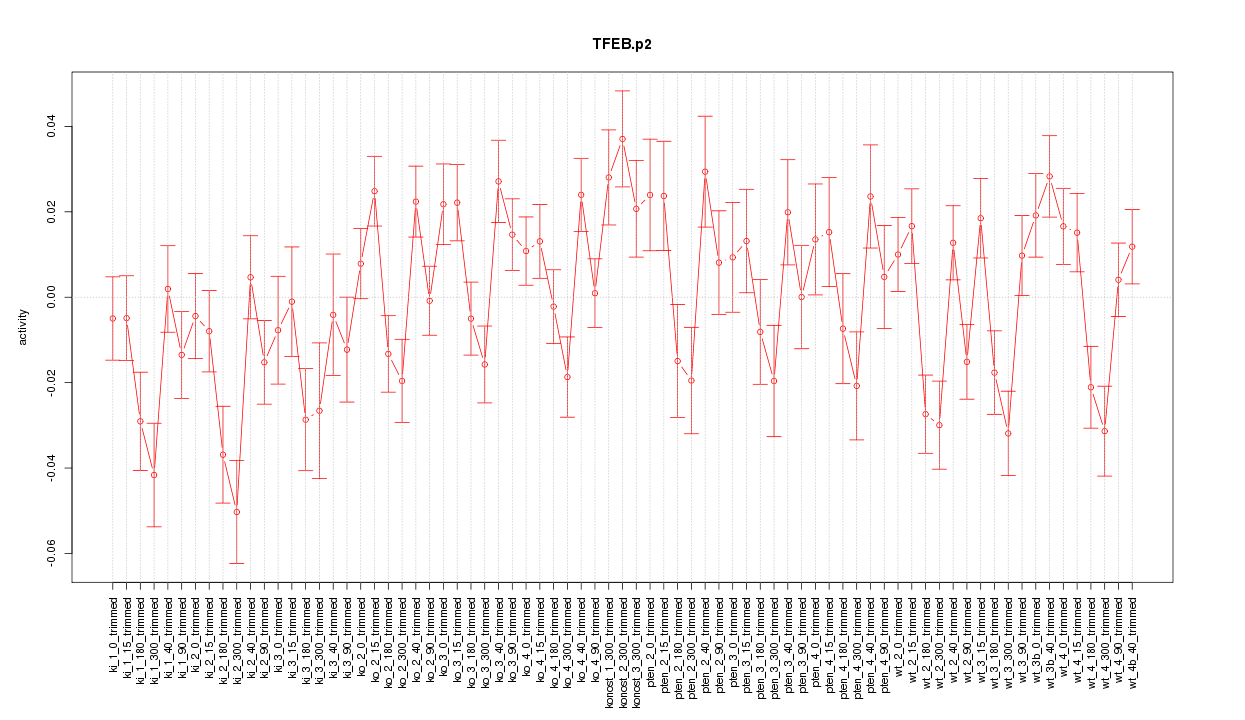
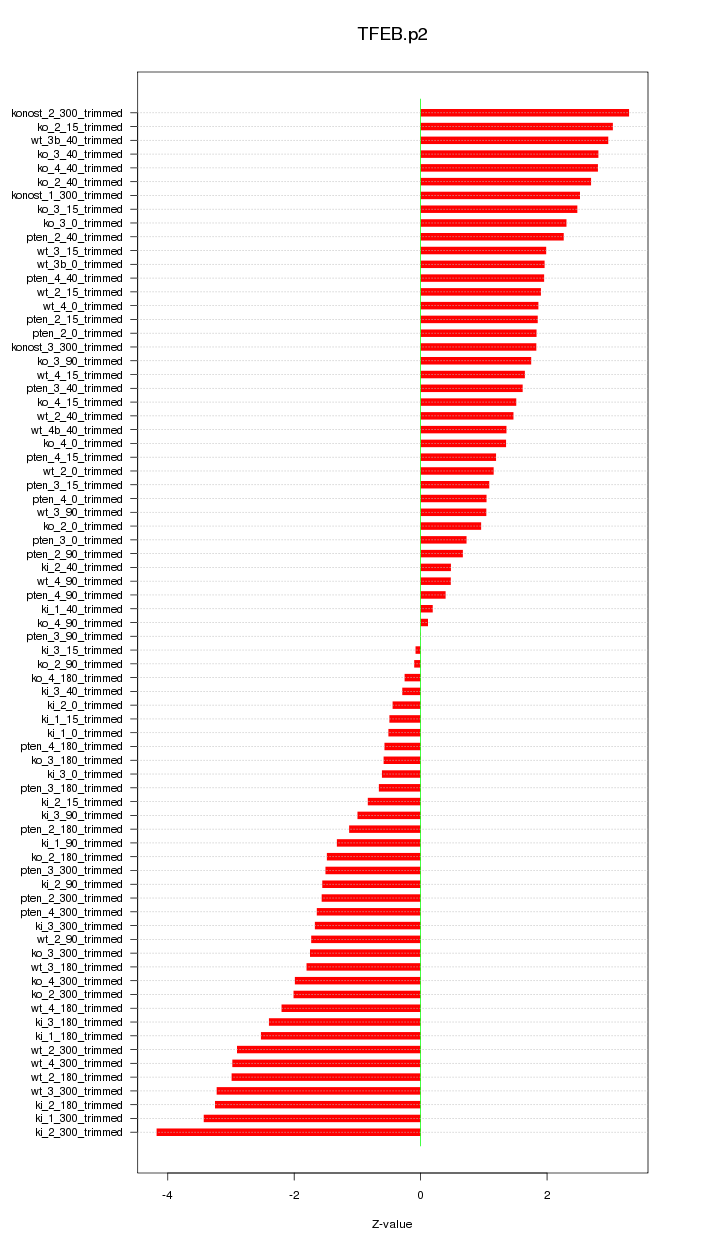
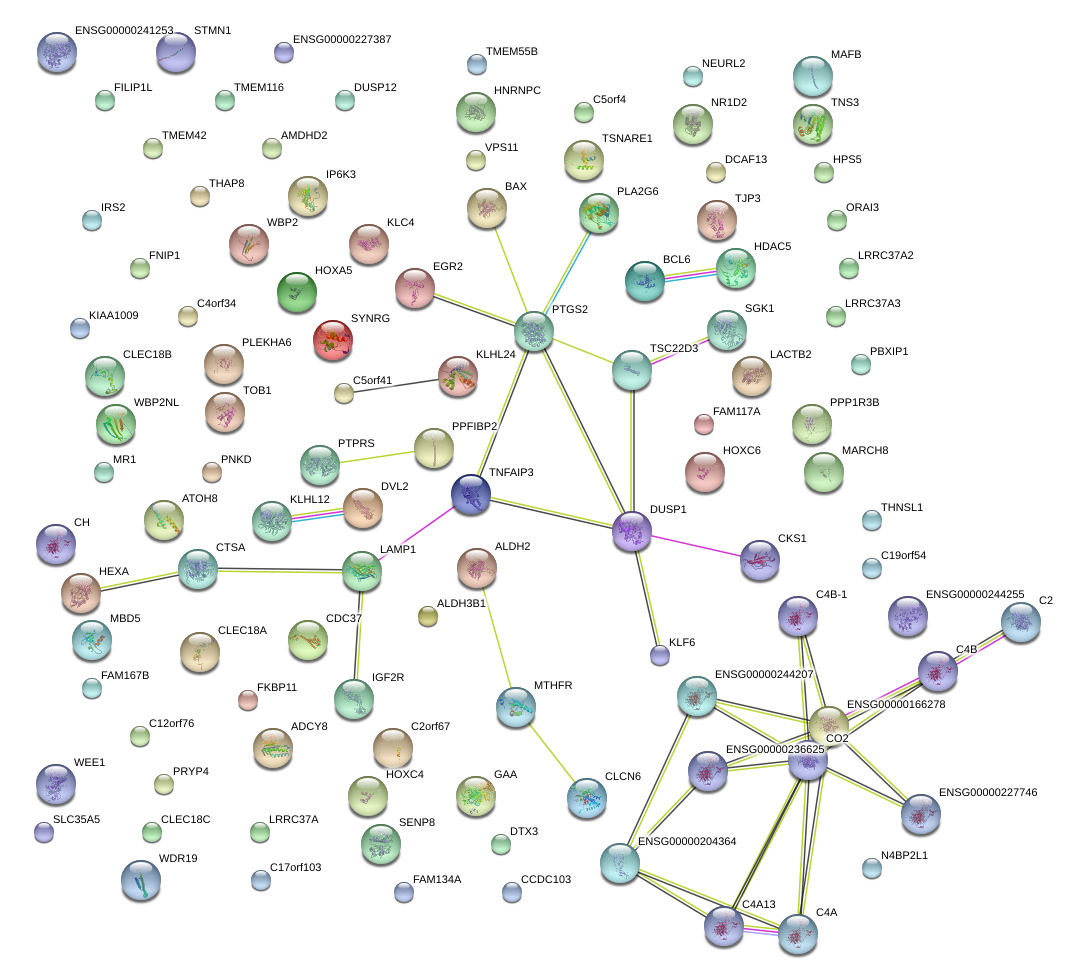
|
chr1_-_11866079
|
31.284
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr5_+_172483285
|
29.576
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_-_11865963
|
25.312
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr19_-_41256349
|
24.596
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr19_-_36545119
|
23.610
|
|
THAP8
|
THAP domain containing 8
|
|
chr19_-_41256371
|
22.878
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr20_-_44519629
|
22.585
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chrX_-_106960268
|
21.634
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_67776016
|
20.742
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr17_-_7137722
|
20.728
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chrX_-_114797017
|
19.474
|
|
|
|
|
chr3_-_187463474
|
18.354
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chrX_-_106960028
|
17.632
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_-_49318663
|
17.122
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr5_-_154230157
|
16.644
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr6_-_33714681
|
16.619
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr5_-_154230212
|
16.558
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr12_+_54447660
|
16.529
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr17_-_7137580
|
15.884
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr15_-_72668301
|
15.742
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_+_49458129
|
15.557
|
|
BAX
|
BCL2-associated X protein
|
|
chr19_-_5340700
|
15.482
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr10_-_64576123
|
15.378
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr17_-_7137675
|
15.187
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr13_-_33002218
|
15.054
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr2_+_85980600
|
14.775
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr3_-_187463227
|
14.736
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_+_138188352
|
14.479
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_+_11866302
|
14.221
|
|
CLCN6
|
chloride channel 6
|
|
chr6_+_160390128
|
14.220
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr19_-_10764449
|
14.053
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr22_-_38577692
|
14.046
|
NM_001004426
NM_003560
NM_001199562
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr1_+_32712817
|
13.834
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr1_+_11866240
|
13.771
|
|
CLCN6
|
chloride channel 6
|
|
chr10_-_64576074
|
13.722
|
|
EGR2
|
early growth response 2
|
|
chr1_-_26232890
|
13.538
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr13_-_110438896
|
13.488
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr11_-_18343670
|
13.480
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr3_+_112280871
|
13.454
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr19_+_49458151
|
13.110
|
|
BAX
|
BCL2-associated X protein
|
|
chr16_+_30960413
|
13.004
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr16_+_30960434
|
12.869
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr3_+_112280911
|
12.851
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr1_-_186649421
|
12.659
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr3_+_23986735
|
12.638
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr16_+_30960377
|
12.583
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr11_-_71814335
|
12.473
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr5_-_154230148
|
12.409
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr15_-_72668184
|
12.362
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr20_-_39317875
|
12.352
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr19_-_36545298
|
12.090
|
|
THAP8
|
THAP domain containing 8
|
|
chr16_+_2570322
|
11.425
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr17_-_42200957
|
11.424
|
|
HDAC5
|
histone deacetylase 5
|
|
chr3_+_183353355
|
11.377
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr5_-_172198160
|
11.320
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr17_-_42201009
|
11.272
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr11_-_71814268
|
11.162
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr11_-_71814294
|
10.952
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr17_-_47841517
|
10.791
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr20_+_44519924
|
10.658
|
|
CTSA
|
cathepsin A
|
|
chr2_+_220042963
|
10.649
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr13_+_113951461
|
10.198
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr12_+_57998600
|
10.186
|
NM_178502
|
DTX3
|
deltex homolog 3 (Drosophila)
|
|
chr19_+_3708358
|
10.133
|
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr7_-_47622127
|
9.875
|
|
TNS3
|
tensin 3
|
|
chr1_-_154928595
|
9.841
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr13_+_52027435
|
9.792
|
|
|
|
|
chr11_-_71814371
|
9.739
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_-_26232643
|
9.672
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr2_+_220042901
|
9.610
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr17_+_78075390
|
9.563
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr15_+_72410630
|
9.378
|
NM_001172111
NM_145204
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr16_+_69984809
|
9.291
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr1_-_154928564
|
9.267
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr3_+_44903376
|
9.239
|
NM_144638
|
TMEM42
|
transmembrane protein 42
|
|
chr17_-_35969401
|
9.091
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr14_-_20929635
|
9.062
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr5_-_131132709
|
9.008
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr3_-_99594915
|
8.951
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr7_-_47621741
|
8.950
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr7_-_27183225
|
8.942
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr11_+_118938473
|
8.939
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_-_47841468
|
8.907
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr22_+_42394728
|
8.832
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr17_-_21156577
|
8.794
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr5_-_131132690
|
8.778
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr2_+_148778573
|
8.746
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr1_-_202896315
|
8.718
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr20_+_44519965
|
8.695
|
|
CTSA
|
cathepsin A
|
|
chr4_-_39640461
|
8.679
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr6_-_84937313
|
8.653
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr17_+_78075360
|
8.598
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr2_-_211035377
|
8.486
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr11_+_1330977
|
8.405
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr17_-_62915580
|
8.388
|
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr19_+_49458031
|
8.366
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr2_+_220042948
|
8.343
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr7_+_27225009
|
8.341
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr9_+_103115342
|
8.224
|
|
|
|
|
chr2_+_220042986
|
8.191
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr1_+_11866206
|
8.170
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr12_-_110505499
|
8.155
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr20_+_44520021
|
8.135
|
|
CTSA
|
cathepsin A
|
|
chr17_-_73851377
|
8.087
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr17_-_48945338
|
8.067
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr6_+_31895401
|
8.064
|
|
C2
|
complement component 2
|
|
chr1_+_181002560
|
7.937
|
NM_001194999
NM_001195000
NM_001195035
NM_001531
|
MR1
|
major histocompatibility complex, class I-related
|
|
chr1_-_202896368
|
7.923
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr1_-_204329043
|
7.896
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr10_+_25305507
|
7.889
|
NM_024838
|
THNSL1
|
threonine synthase-like 1 (S. cerevisiae)
|
|
chr3_+_23987514
|
7.764
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr11_+_7535213
|
7.736
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr11_+_118938528
|
7.585
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr15_-_72668740
|
7.585
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr10_-_46090191
|
7.575
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr8_-_9009050
|
7.474
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr14_-_20929484
|
7.472
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr5_-_131132642
|
7.414
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr8_-_143484528
|
7.342
|
NM_145003
|
TSNARE1
|
t-SNARE domain containing 1
|
|
chr6_-_134497069
|
7.320
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_+_118938515
|
7.310
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_+_42977141
|
7.285
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|
|
chr4_+_39184023
|
7.244
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chr8_-_71581354
|
7.197
|
|
LACTB2
|
lactamase, beta 2
|
|
chr13_+_113951537
|
7.191
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr11_+_9595309
|
7.170
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr6_+_43027368
|
7.073
|
NM_138343
NM_201521
|
KLC4
|
kinesin light chain 4
|
|
chr12_-_112450914
|
6.922
|
NM_001193453
NM_001193531
NM_138341
|
LOC728543
TMEM116
|
uncharacterized LOC728543
transmembrane protein 116
|
|
chr10_-_3827418
|
6.832
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr11_+_118938530
|
6.811
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr6_+_31982538
|
6.761
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr11_+_9595183
|
6.736
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr14_-_92572950
|
6.717
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr19_+_14544100
|
6.716
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr14_+_103388992
|
6.713
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr3_-_4508952
|
6.688
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr5_-_39219578
|
6.669
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chrX_+_101975641
|
6.648
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr9_+_6757919
|
6.646
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr16_-_28503079
|
6.638
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr3_-_4508955
|
6.590
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr13_+_113951589
|
6.584
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr3_-_4508928
|
6.582
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_+_113465860
|
6.554
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr1_-_154192882
|
6.505
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr4_-_23891608
|
6.492
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr4_+_77135192
|
6.489
|
NM_001242936
|
FAM47E
|
family with sequence similarity 47, member E
|
|
chr8_+_142402108
|
6.476
|
|
|
|
|
chr6_+_31949800
|
6.458
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr22_+_35695872
|
6.443
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr3_-_4508939
|
6.442
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr11_+_9595451
|
6.404
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr15_+_72410724
|
6.397
|
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr5_-_131132715
|
6.394
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr19_+_40854529
|
6.382
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr5_-_139944045
|
6.379
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr10_+_99344421
|
6.355
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr10_+_89102474
|
6.281
|
|
LOC677759
|
uncharacterized LOC677759
|
|
chr2_+_187558654
|
6.264
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr15_-_34629941
|
6.237
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_-_65153148
|
6.227
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr18_+_44497512
|
6.193
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chrX_+_102883891
|
6.139
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr1_+_183605207
|
6.130
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chrX_+_10124984
|
6.121
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chrX_-_14047958
|
6.116
|
NM_001042479
NM_001042480
NM_017856
|
GEMIN8
|
gem (nuclear organelle) associated protein 8
|
|
chr8_-_95961543
|
6.086
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr1_-_24151932
|
6.076
|
NM_000191
NM_001166059
|
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase
|
|
chr19_+_50380681
|
6.013
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr1_-_155211049
|
5.962
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr11_+_9595429
|
5.954
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr12_-_65153055
|
5.933
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr7_-_47621635
|
5.919
|
|
TNS3
|
tensin 3
|
|
chr3_+_113465901
|
5.916
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr7_+_36429388
|
5.829
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr12_+_96337070
|
5.806
|
NM_152435
|
AMDHD1
|
amidohydrolase domain containing 1
|
|
chr14_-_64970319
|
5.800
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr10_-_3827205
|
5.790
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr3_+_113465910
|
5.790
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr2_-_172967627
|
5.759
|
|
DLX2
|
distal-less homeobox 2
|
|
chr4_-_149363498
|
5.680
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr8_-_71581419
|
5.635
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr9_-_140444835
|
5.634
|
NM_001098537
NM_152286
|
PNPLA7
|
patatin-like phospholipase domain containing 7
|
|
chr17_+_79935493
|
5.613
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr19_-_40324747
|
5.605
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr7_+_27224163
|
5.575
|
|
|
|
|
chr6_+_123110345
|
5.557
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr12_-_65153182
|
5.544
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr21_-_36421558
|
5.500
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_+_48172508
|
5.485
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr1_-_24151845
|
5.479
|
|
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase
|
|
chr7_+_99070513
|
5.456
|
NM_001013258
NM_213603
|
ZNF789
|
zinc finger protein 789
|
|
chr9_-_123476578
|
5.444
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr4_-_103266577
|
5.341
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr11_-_66335958
|
5.328
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr19_+_14544216
|
5.323
|
|
PKN1
|
protein kinase N1
|
|
chr20_-_2821120
|
5.322
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr3_-_40350948
|
5.236
|
|
FLJ33065
|
uncharacterized LOC440952
|
|
chr7_+_99775521
|
5.181
|
NM_012447
|
STAG3
|
stromal antigen 3
|
|
chr1_+_10092990
|
5.174
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|