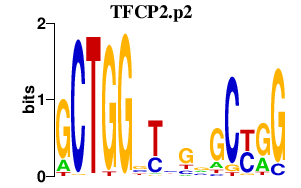
|
chr17_+_74380676
|
7.493
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr2_-_46384
|
6.635
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr20_+_43803539
|
6.600
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr19_-_51487070
|
5.700
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr17_+_74380943
|
5.484
|
|
SPHK1
|
sphingosine kinase 1
|
|
chrX_-_110513750
|
5.452
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr13_-_36705432
|
5.334
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr11_-_2019070
|
5.242
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr13_-_36705513
|
5.117
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr20_-_31172874
|
5.056
|
|
LOC284804
|
uncharacterized LOC284804
|
|
chr8_-_143867931
|
5.048
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr17_+_21187967
|
4.940
|
NM_145109
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr17_-_43510204
|
4.793
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr1_-_1475739
|
4.757
|
NM_001114748
|
TMEM240
|
transmembrane protein 240
|
|
chr11_+_65082288
|
4.745
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr17_+_21188021
|
4.706
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr17_+_21188032
|
4.674
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr4_+_4861371
|
4.567
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr17_-_39661803
|
4.544
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr1_-_41328019
|
4.514
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr15_+_32322690
|
4.425
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr19_-_1567875
|
4.354
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr4_-_57522059
|
4.319
|
|
HOPX
|
HOP homeobox
|
|
chr1_-_16482426
|
4.278
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr1_-_205912546
|
4.258
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr20_-_62462568
|
4.143
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr4_+_84457390
|
4.092
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr2_-_73520448
|
4.027
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr3_-_45267620
|
3.988
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr1_-_935352
|
3.944
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr19_-_43969723
|
3.929
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr17_-_80797889
|
3.904
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr3_-_52090090
|
3.821
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr1_-_935469
|
3.775
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr17_-_43510279
|
3.628
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr11_-_2019007
|
3.549
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_-_1182101
|
3.501
|
NM_001014980
|
FAM132A
|
family with sequence similarity 132, member A
|
|
chr1_-_22469414
|
3.470
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr2_-_96804173
|
3.422
|
NM_001002036
|
ASTL
|
astacin-like metallo-endopeptidase (M12 family)
|
|
chr7_-_73133917
|
3.409
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr17_-_39222126
|
3.404
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr7_+_28448970
|
3.387
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr13_-_36705466
|
3.344
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr12_-_45270632
|
3.334
|
NM_001145107
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr19_+_15726028
|
3.253
|
NM_007253
|
CYP4F8
|
cytochrome P450, family 4, subfamily F, polypeptide 8
|
|
chr19_-_1237840
|
3.228
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr19_+_49055421
|
3.225
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr10_-_88126201
|
3.217
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr14_-_71275732
|
3.205
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr17_-_78450247
|
3.180
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr7_-_559030
|
3.174
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr7_-_558810
|
3.134
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr1_-_36851517
|
3.124
|
|
STK40
|
serine/threonine kinase 40
|
|
chr4_+_84457256
|
3.084
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr1_-_41328002
|
3.069
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr1_-_36851484
|
3.068
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr1_-_166135978
|
3.061
|
|
|
|
|
chr14_+_79745653
|
3.012
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chrX_-_17879355
|
2.996
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr11_+_67810446
|
2.968
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr17_-_34079896
|
2.960
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr17_-_17399494
|
2.937
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr4_+_8200918
|
2.933
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr15_-_74495093
|
2.897
|
NM_001199042
NM_022369
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr12_-_45269340
|
2.887
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr20_-_62168663
|
2.881
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr15_+_81071708
|
2.840
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chrX_+_152770083
|
2.827
|
|
BGN
|
biglycan
|
|
chr11_-_6790187
|
2.807
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chrX_+_68725077
|
2.799
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr19_+_33685140
|
2.797
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr2_+_217498205
|
2.739
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr1_-_156675237
|
2.722
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr5_-_134914447
|
2.713
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr12_-_53207843
|
2.685
|
NM_002272
|
KRT4
|
keratin 4
|
|
chr5_-_73936299
|
2.681
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr12_-_105501410
|
2.674
|
|
LOC414300
|
uncharacterized LOC414300
|
|
chr19_+_15751706
|
2.672
|
NM_000896
NM_001199208
NM_001199209
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr1_+_229761962
|
2.671
|
NM_014777
|
URB2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr20_+_51589752
|
2.669
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr19_-_55895602
|
2.652
|
NM_001190764
|
TMEM238
|
transmembrane protein 238
|
|
chr2_+_217498082
|
2.572
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr19_+_1207129
|
2.572
|
|
EFNA2
|
ephrin-A2
|
|
chrX_-_110513665
|
2.561
|
|
CAPN6
|
calpain 6
|
|
chr15_-_42264687
|
2.555
|
|
EHD4
|
EH-domain containing 4
|
|
chr22_+_31478845
|
2.551
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chrX_+_152760346
|
2.549
|
NM_001711
|
BGN
|
biglycan
|
|
chr1_+_229761988
|
2.529
|
|
URB2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr20_-_62711160
|
2.524
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr11_-_123612369
|
2.513
|
|
ZNF202
|
zinc finger protein 202
|
|
chr1_+_90098624
|
2.512
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr13_+_37006628
|
2.501
|
|
CCNA1
|
cyclin A1
|
|
chr19_-_33793321
|
2.498
|
|
|
|
|
chr15_-_74495528
|
2.495
|
NM_001142618
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr7_+_48128225
|
2.485
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr7_-_44365019
|
2.478
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr1_-_31230594
|
2.465
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr1_+_36621457
|
2.464
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr1_+_39456887
|
2.444
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr10_+_5566923
|
2.438
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr6_+_160148485
|
2.431
|
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr19_-_45737466
|
2.427
|
NM_138568
|
EXOC3L2
|
exocyst complex component 3-like 2
|
|
chr17_+_56065408
|
2.412
|
|
|
|
|
chr17_+_81037504
|
2.403
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr20_+_361938
|
2.402
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr17_+_30814708
|
2.402
|
|
|
|
|
chr3_+_128199744
|
2.393
|
|
|
|
|
chr6_+_35310317
|
2.388
|
NM_001171818
NM_001171819
NM_001171820
NM_006238
NM_177435
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr16_+_84853585
|
2.382
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr11_+_46366816
|
2.381
|
NM_201533
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr8_-_143823823
|
2.374
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr16_+_577833
|
2.361
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr9_-_120177307
|
2.348
|
NM_014010
|
ASTN2
|
astrotactin 2
|
|
chr4_-_57522469
|
2.329
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr6_+_126070895
|
2.328
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr9_-_140335790
|
2.328
|
NM_001033113
NM_198585
|
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8
|
|
chr19_+_15783827
|
2.324
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr17_+_30814552
|
2.319
|
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr22_-_42322778
|
2.310
|
NM_052945
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C
|
|
chr13_+_37006408
|
2.306
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr15_-_101792125
|
2.294
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr11_-_2018885
|
2.292
|
|
H19
RPS12
|
H19, imprinted maternally expressed transcript (non-protein coding)
ribosomal protein S12
|
|
chr5_+_175792444
|
2.283
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr18_+_48086402
|
2.281
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr17_-_73874634
|
2.271
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr20_+_19193282
|
2.266
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr1_-_19282773
|
2.262
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr1_-_31230673
|
2.259
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr11_-_2159784
|
2.256
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_+_77648127
|
2.254
|
|
TMEM63C
|
transmembrane protein 63C
|
|
chr19_+_14142572
|
2.252
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr22_+_38035683
|
2.249
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr3_-_10547267
|
2.247
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr11_+_65779461
|
2.242
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr18_+_47087057
|
2.239
|
|
LIPG
|
lipase, endothelial
|
|
chr1_-_228135587
|
2.230
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr6_+_35310368
|
2.227
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr2_-_110874142
|
2.195
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr8_+_144680073
|
2.193
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr11_+_46366982
|
2.193
|
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr2_-_96810775
|
2.192
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr11_+_65554566
|
2.188
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr17_+_4402128
|
2.175
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr17_+_635656
|
2.171
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chrX_-_71526583
|
2.167
|
NM_004143
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr3_-_50649110
|
2.162
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr10_+_6244839
|
2.140
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr17_-_39675078
|
2.138
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr11_-_2018703
|
2.137
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr9_+_35605275
|
2.121
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr5_-_139725860
|
2.105
|
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr7_+_143078447
|
2.104
|
|
ZYX
|
zyxin
|
|
chr11_+_130029699
|
2.093
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr6_+_15245702
|
2.092
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_+_176957448
|
2.092
|
NM_000523
|
HOXD13
|
homeobox D13
|
|
chr19_+_45754505
|
2.090
|
NM_001199867
NM_031417
|
MARK4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr16_+_67465019
|
2.086
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr3_+_5021141
|
2.085
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr2_-_43453724
|
2.073
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr6_+_111195986
|
2.064
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr14_-_105767274
|
2.056
|
|
BRF1
|
BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae)
|
|
chr17_+_44928947
|
2.046
|
NM_003396
|
WNT9B
|
wingless-type MMTV integration site family, member 9B
|
|
chr11_+_129685732
|
2.033
|
NM_138788
|
TMEM45B
|
transmembrane protein 45B
|
|
chr1_+_90098693
|
2.032
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr9_+_92219926
|
2.014
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr12_+_132414240
|
2.013
|
|
|
|
|
chr2_+_242295660
|
2.010
|
NM_014808
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2
|
|
chr9_+_35605339
|
2.000
|
|
TESK1
|
testis-specific kinase 1
|
|
chr8_-_139509064
|
1.986
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr12_-_55042135
|
1.980
|
NM_053283
|
DCD
|
dermcidin
|
|
chr4_-_164394885
|
1.970
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr11_+_1855673
|
1.944
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr1_-_95392501
|
1.935
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr20_+_35089817
|
1.932
|
NM_001042486
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr8_-_26722915
|
1.918
|
NM_000680
NM_033302
NM_033303
NM_033304
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr1_+_89990396
|
1.915
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr15_+_31619043
|
1.914
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr22_+_45705766
|
1.904
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr1_+_90098643
|
1.900
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr11_+_94277566
|
1.900
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr22_+_32439018
|
1.895
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr20_-_23030141
|
1.893
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr19_+_14142611
|
1.893
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr11_-_66084501
|
1.884
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr6_+_31496738
|
1.873
|
NM_001011700
|
MCCD1
|
mitochondrial coiled-coil domain 1
|
|
chr9_+_140125208
|
1.871
|
NM_001177316
NM_001177317
|
SLC34A3
|
solute carrier family 34 (sodium phosphate), member 3
|
|
chr7_-_30739660
|
1.863
|
NM_001202475
NM_001202481
|
CRHR2
|
corticotropin releasing hormone receptor 2
|
|
chr19_-_10047014
|
1.862
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chrX_+_152760452
|
1.858
|
|
BGN
|
biglycan
|
|
chr3_-_128206704
|
1.857
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr7_-_99756275
|
1.854
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr11_+_130318457
|
1.846
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr11_+_46354454
|
1.829
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr16_-_2908105
|
1.823
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr22_+_31487315
|
1.822
|
|
SMTN
|
smoothelin
|
|
chr11_+_130029712
|
1.820
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr5_-_176924552
|
1.819
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr15_-_101791674
|
1.818
|
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr10_-_103535656
|
1.818
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr1_-_166135951
|
1.816
|
NM_001017961
|
FAM78B
|
family with sequence similarity 78, member B
|