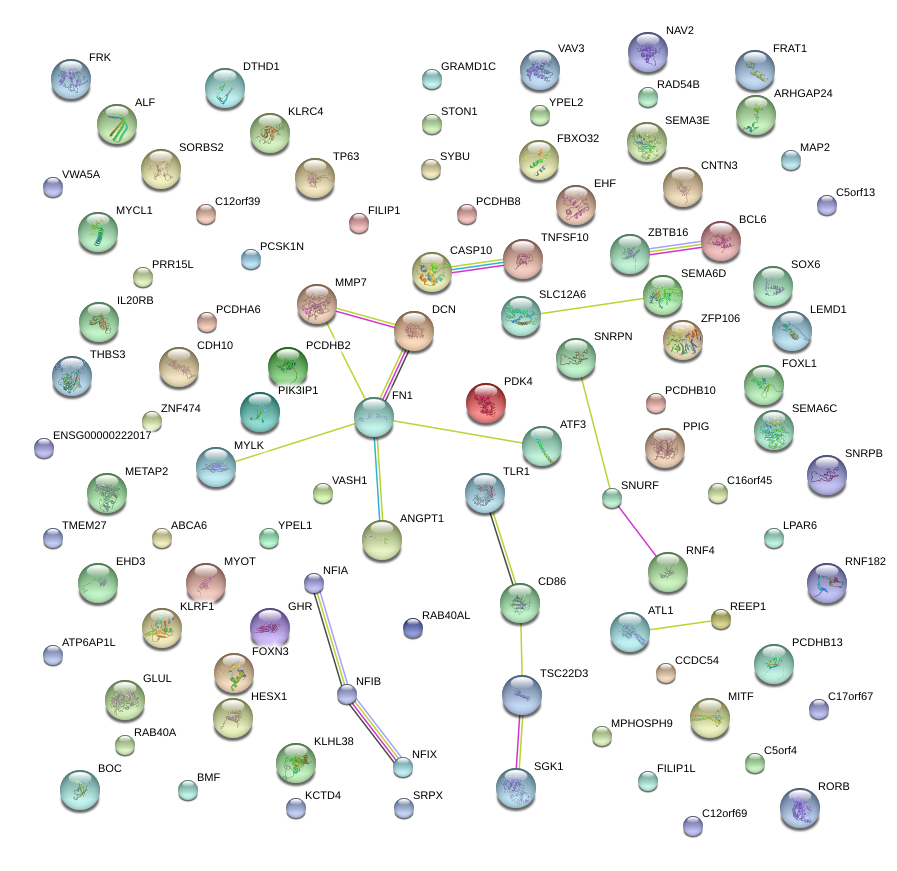
|
chr1_-_108231122
|
12.486
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr22_-_31688329
|
12.476
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr2_-_216246961
|
12.006
|
|
FN1
|
fibronectin 1
|
|
chr2_-_216245573
|
11.778
|
|
FN1
|
fibronectin 1
|
|
chr7_-_83270746
|
10.485
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr2_-_216242916
|
10.073
|
|
FN1
|
fibronectin 1
|
|
chr5_+_140474198
|
9.920
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr12_-_91573264
|
8.200
|
NM_133503
|
DCN
|
decorin
|
|
chr13_-_45775174
|
7.734
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr4_-_186732047
|
7.682
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_-_102401430
|
7.372
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr15_-_40401041
|
6.952
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr4_-_186732208
|
6.835
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_140247767
|
6.678
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr1_-_40367539
|
6.579
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr6_-_134498973
|
6.565
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_-_216259197
|
6.467
|
|
FN1
|
fibronectin 1
|
|
chr8_-_108510094
|
6.440
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr11_-_16424391
|
6.405
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr5_-_24645084
|
6.301
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr5_+_137203559
|
6.247
|
|
MYOT
|
myotilin
|
|
chr3_+_121774208
|
6.246
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr2_-_216289936
|
6.230
|
|
FN1
|
fibronectin 1
|
|
chr11_+_34654010
|
5.797
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr7_-_142985270
|
5.746
|
|
|
|
|
chr5_+_81601165
|
5.731
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr18_+_3594056
|
5.617
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr15_+_48010685
|
5.605
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr15_+_25200069
|
5.577
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr12_-_14967091
|
5.533
|
NM_001013698
|
C12orf69
|
chromosome 12 open reading frame 69
|
|
chr12_-_10562142
|
5.362
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 readthrough
killer cell lectin-like receptor subfamily C, member 4
|
|
chr5_+_137203544
|
5.223
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr4_-_186697065
|
5.201
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_-_89883306
|
5.144
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr3_-_74570262
|
5.079
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr6_-_116381765
|
5.076
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr1_-_182357757
|
5.053
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_-_123339178
|
5.050
|
|
MYLK
|
myosin light chain kinase
|
|
chr14_+_77228940
|
5.022
|
|
VASH1
|
vasohibin 1
|
|
chr2_+_210444364
|
4.986
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr6_+_13925423
|
4.983
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr17_-_67138013
|
4.895
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr3_-_187454247
|
4.882
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr9_+_127420690
|
4.868
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr11_+_123986103
|
4.860
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr12_+_9980076
|
4.800
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr7_-_95225802
|
4.733
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr15_-_34610928
|
4.686
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr14_+_51026742
|
4.570
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr1_+_212788359
|
4.554
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr3_-_99833348
|
4.548
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr16_+_86612114
|
4.494
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr1_-_182357871
|
4.479
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chrX_-_102774416
|
4.473
|
NM_080879
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chrX_-_15683153
|
4.443
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr3_+_113616317
|
4.320
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr8_-_124665165
|
4.312
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr5_+_140593508
|
4.281
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr3_+_69915374
|
4.274
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr17_-_46035109
|
4.263
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr4_-_38806410
|
4.232
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr8_-_124553253
|
4.226
|
|
FBXO32
|
F-box protein 32
|
|
chr5_+_121465214
|
4.179
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr4_+_86699858
|
4.178
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr10_+_99079017
|
4.151
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr6_-_134639195
|
3.991
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_202047803
|
3.833
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr4_-_186578122
|
3.779
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr13_-_48987017
|
3.700
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr2_-_216249682
|
3.688
|
|
FN1
|
fibronectin 1
|
|
chr2_-_56412904
|
3.668
|
|
LOC100129434
|
uncharacterized LOC100129434
|
|
chr16_+_15528324
|
3.664
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr2_+_48757292
|
3.657
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr3_+_113557679
|
3.624
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr4_-_186877413
|
3.611
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_-_95449131
|
3.601
|
NM_001205263
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chrX_-_106959597
|
3.569
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_+_21679190
|
3.565
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr6_-_76203256
|
3.550
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr2_-_86564632
|
3.538
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr8_-_110655418
|
3.528
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_+_13925097
|
3.515
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chrX_-_38079980
|
3.493
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr5_-_121814781
|
3.445
|
|
|
|
|
chr9_-_14314036
|
3.438
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_57234279
|
3.436
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr11_+_20049128
|
3.432
|
|
NAV2
|
neuron navigator 2
|
|
chr5_+_42565562
|
3.406
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr9_+_77112251
|
3.403
|
NM_006914
|
RORB
|
RAR-related orphan receptor B
|
|
chr5_+_140557370
|
3.362
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr3_-_172241060
|
3.357
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr12_-_123706393
|
3.352
|
NM_022782
|
MPHOSPH9
|
M-phase phosphoprotein 9
|
|
chr17_-_54893249
|
3.330
|
NM_001085430
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr4_-_186733372
|
3.312
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_48693934
|
3.299
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr3_+_107096187
|
3.298
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr5_+_42549657
|
3.293
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr17_+_57408907
|
3.269
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr2_+_31456879
|
3.247
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr1_-_205391180
|
3.216
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr13_-_48987247
|
3.212
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr3_+_69812961
|
3.200
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_36285643
|
3.200
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr1_-_151118932
|
3.197
|
NM_001178061
NM_001178062
NM_030913
|
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C
|
|
chr3_+_112931374
|
3.181
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr3_+_136676706
|
3.167
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr4_-_186577858
|
3.156
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_154230157
|
3.149
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr5_-_111312522
|
3.147
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_7212868
|
3.144
|
|
FAM66B
|
family with sequence similarity 66, member B
|
|
chr5_+_140566655
|
3.143
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr5_-_154230212
|
3.128
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr2_-_201967702
|
3.123
|
|
|
|
|
chr3_-_99594915
|
3.100
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_182360181
|
3.051
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_42550181
|
3.048
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr3_+_189507448
|
3.047
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr11_+_113931228
|
3.021
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr3_+_69812620
|
3.014
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_-_155177685
|
3.011
|
NM_001252607
NM_001252608
NM_007112
|
THBS3
|
thrombospondin 3
|
|
chr6_-_2751153
|
2.990
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr4_+_71384289
|
2.985
|
NM_212557
|
AMTN
|
amelotin
|
|
chr1_-_183387404
|
2.961
|
|
|
|
|
chr5_-_111091947
|
2.950
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr13_+_43136871
|
2.907
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr19_-_14939275
|
2.873
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr1_+_153940345
|
2.865
|
NM_130898
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4
|
|
chr4_-_105416050
|
2.829
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr1_+_201476283
|
2.821
|
|
|
|
|
chr1_+_116654375
|
2.813
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr12_-_91572328
|
2.796
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr8_-_57123858
|
2.791
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr4_-_110723127
|
2.777
|
|
CFI
|
complement factor I
|
|
chr9_-_4666665
|
2.776
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr3_-_93747453
|
2.772
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr20_+_10415949
|
2.768
|
NM_001009608
|
C20orf94
|
chromosome 20 open reading frame 94
|
|
chr9_-_36276930
|
2.758
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr1_+_84768333
|
2.747
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_-_137071703
|
2.746
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr9_+_90112754
|
2.744
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr9_-_14086901
|
2.738
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_87012758
|
2.738
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr2_+_202047620
|
2.732
|
NM_001206524
NM_001206542
NM_001230
NM_032974
NM_032976
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr9_-_89562103
|
2.719
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr14_-_92413738
|
2.717
|
|
FBLN5
|
fibulin 5
|
|
chr3_-_12586962
|
2.715
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr8_-_133772808
|
2.713
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr5_+_35856990
|
2.701
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr5_+_71502700
|
2.680
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr14_-_21493122
|
2.679
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr2_+_38177595
|
2.646
|
NM_001170793
NM_144713
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr2_-_225811730
|
2.641
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr11_+_27015627
|
2.637
|
NM_203371
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr2_+_30454396
|
2.636
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr17_-_40264687
|
2.634
|
NM_024119
|
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr11_+_19372270
|
2.631
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr12_-_92539614
|
2.628
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr13_-_33780142
|
2.622
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_163099877
|
2.616
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr15_+_80696569
|
2.585
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr12_+_32654976
|
2.569
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_-_158390266
|
2.566
|
NM_020169
|
LXN
|
latexin
|
|
chr11_+_118401873
|
2.562
|
|
TMEM25
|
transmembrane protein 25
|
|
chr11_+_22696381
|
2.559
|
|
GAS2
|
growth arrest-specific 2
|
|
chr3_-_105421044
|
2.546
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr10_+_17271276
|
2.526
|
|
VIM
|
vimentin
|
|
chr5_-_154230148
|
2.508
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr7_-_107643330
|
2.506
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr10_+_17271302
|
2.497
|
|
VIM
|
vimentin
|
|
chr19_-_57988891
|
2.495
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr4_+_39063851
|
2.493
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr13_+_53035227
|
2.445
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr1_+_164528914
|
2.433
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_-_26277807
|
2.432
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr13_-_48987652
|
2.429
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr9_+_123884043
|
2.429
|
|
CNTRL
|
centriolin
|
|
chr15_-_60919642
|
2.424
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_+_79439432
|
2.423
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr8_-_124553448
|
2.420
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chrX_+_84498993
|
2.418
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr10_-_105845619
|
2.410
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr6_+_139456240
|
2.398
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr10_+_95848811
|
2.395
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr9_-_98268866
|
2.390
|
|
PTCH1
|
patched 1
|
|
chr7_-_7575449
|
2.387
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr4_+_86699846
|
2.356
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr10_+_17271277
|
2.353
|
|
VIM
|
vimentin
|
|
chr11_+_20044362
|
2.344
|
|
NAV2
|
neuron navigator 2
|
|
chr12_+_14538232
|
2.337
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr10_-_105845556
|
2.301
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr1_+_33722158
|
2.289
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr20_+_33134691
|
2.270
|
NM_181509
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr7_-_82792127
|
2.263
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr6_-_49712055
|
2.259
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr2_-_192711923
|
2.248
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr14_-_23652840
|
2.246
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr13_-_33760215
|
2.233
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_195310770
|
2.216
|
|
APOD
|
apolipoprotein D
|
|
chr2_+_201450579
|
2.210
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr2_+_128175989
|
2.204
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|