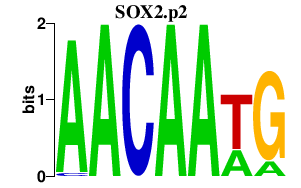
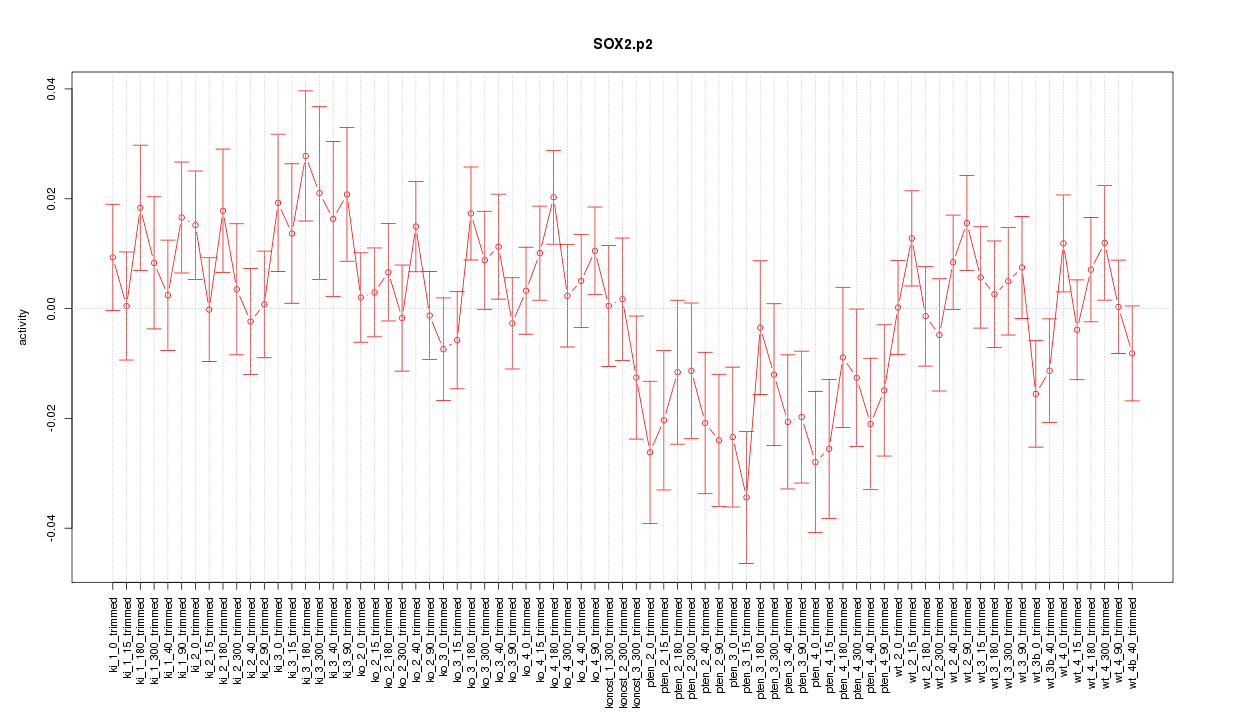
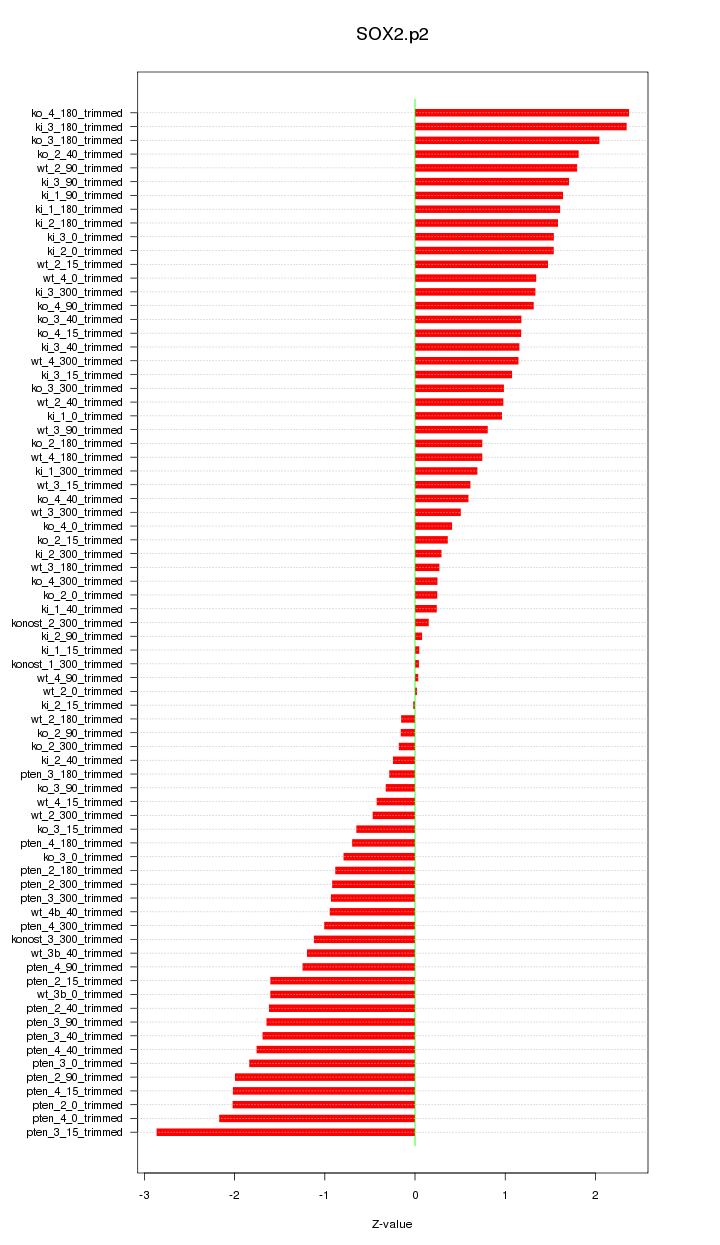
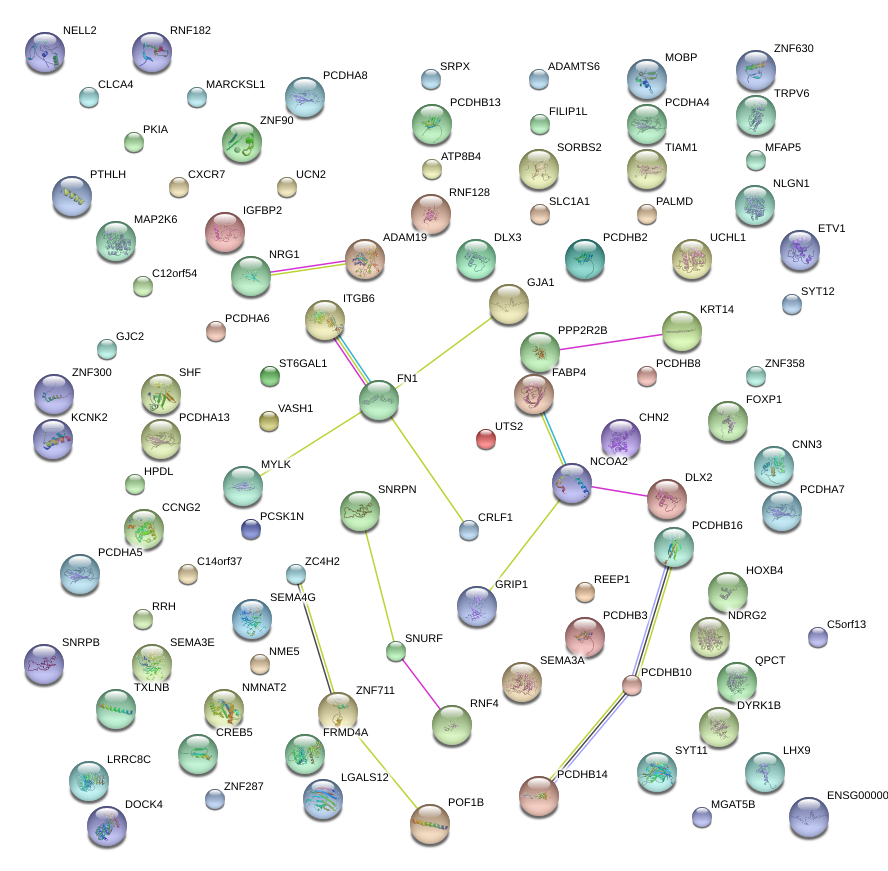
|
chr5_+_140474198
|
28.294
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr15_+_25200069
|
27.070
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr4_+_41258941
|
15.718
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr5_+_140254930
|
14.460
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chrX_+_84499037
|
14.268
|
|
ZNF711
|
zinc finger protein 711
|
|
chr14_+_77228940
|
14.243
|
|
VASH1
|
vasohibin 1
|
|
chrX_-_64196268
|
12.361
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr5_+_140235594
|
11.983
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr5_+_140201221
|
11.848
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr12_-_28124915
|
11.288
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr17_-_16472463
|
11.255
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr3_+_186648515
|
10.701
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr10_+_102732285
|
9.994
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr5_-_150284293
|
9.943
|
|
ZNF300
|
zinc finger protein 300
|
|
chr3_+_186648273
|
9.742
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chrX_+_84498993
|
9.437
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr5_-_150284503
|
9.285
|
NM_001172831
NM_001172832
NM_052860
|
ZNF300
|
zinc finger protein 300
|
|
chrX_-_38079980
|
9.024
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr5_+_140480056
|
8.958
|
NM_018937
|
PCDHB3
|
protocadherin beta 3
|
|
chr4_+_41258909
|
8.939
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr12_-_8815266
|
8.879
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr5_-_146435589
|
8.781
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr2_-_201967702
|
8.719
|
|
|
|
|
chr2_-_161056762
|
8.718
|
|
ITGB6
|
integrin, beta 6
|
|
chr4_-_186877413
|
8.711
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732047
|
8.706
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_84634664
|
8.554
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr17_-_48072566
|
8.204
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chrX_-_84634742
|
7.975
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr3_+_173116243
|
7.513
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr12_-_45307710
|
7.480
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_+_215256559
|
7.478
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_+_100111430
|
7.404
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chrX_-_84634396
|
7.378
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr17_-_39743085
|
7.146
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr5_-_157002739
|
7.064
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr3_-_99594915
|
7.044
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_-_8815370
|
6.959
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr4_+_41258895
|
6.868
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr2_-_86564632
|
6.815
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr7_+_28452143
|
6.744
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_186732208
|
6.707
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_228337414
|
6.685
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr6_+_13925423
|
6.589
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr12_-_8815479
|
6.560
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr5_-_150284429
|
6.529
|
|
ZNF300
|
zinc finger protein 300
|
|
chr5_-_137475076
|
6.405
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr5_-_150284390
|
6.384
|
|
ZNF300
|
zinc finger protein 300
|
|
chr7_-_83824216
|
6.304
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr9_+_4490193
|
6.239
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr5_+_140593508
|
6.136
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr15_-_45493372
|
6.025
|
NM_138356
|
SHF
|
Src homology 2 domain containing F
|
|
chr3_-_71353910
|
6.004
|
NM_001244812
|
FOXP1
|
forkhead box P1
|
|
chr5_+_140777603
|
5.854
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr21_-_32931289
|
5.770
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr6_+_21666645
|
5.682
|
|
LINC00340
|
long intergenic non-protein coding RNA 340
|
|
chrX_-_47931000
|
5.538
|
NM_001190255
|
ZNF630
|
zinc finger protein 630
|
|
chr12_+_48876285
|
5.475
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr2_+_217498082
|
5.417
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr7_-_83270746
|
5.349
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr7_-_111846384
|
5.318
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr17_+_74864427
|
5.116
|
NM_001199172
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr17_-_46655544
|
5.107
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr5_+_140207562
|
4.977
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr8_-_82395401
|
4.946
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr5_+_140213968
|
4.871
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr5_+_140602914
|
4.866
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr19_+_7580987
|
4.856
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr3_-_99833348
|
4.805
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr5_+_140186658
|
4.764
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr1_+_45792544
|
4.734
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr11_+_66790189
|
4.694
|
NM_001177880
|
SYT12
|
synaptotagmin XII
|
|
chr7_+_28725597
|
4.676
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_+_87012758
|
4.642
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr12_-_67072881
|
4.622
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr3_-_48601200
|
4.619
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr5_+_140560979
|
4.611
|
NM_020957
|
PCDHB16
|
protocadherin beta 16
|
|
chr1_-_95392501
|
4.601
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr12_-_67072752
|
4.600
|
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr14_-_21493122
|
4.586
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr5_-_157002705
|
4.580
|
|
|
|
|
chr19_+_19976625
|
4.555
|
NM_021047
|
ZNF253
|
zinc finger protein 253
|
|
chr2_-_216246961
|
4.467
|
|
FN1
|
fibronectin 1
|
|
chr1_-_32801606
|
4.459
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr19_-_40324747
|
4.444
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr5_-_146460991
|
4.388
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr17_+_67410825
|
4.378
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr7_-_142583260
|
4.373
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr1_-_183387404
|
4.333
|
|
|
|
|
chr1_+_197886499
|
4.265
|
NM_020204
|
LHX9
|
LIM homeobox 9
|
|
chr19_-_18717577
|
4.210
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr12_-_45270071
|
4.152
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr4_+_78078394
|
4.125
|
|
CCNG2
|
cyclin G2
|
|
chr19_-_18717631
|
4.124
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr8_+_79428335
|
4.113
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chrX_-_48693934
|
4.108
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr5_+_140557370
|
4.088
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr2_+_37571752
|
4.026
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr17_+_67498391
|
4.017
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr12_-_28122884
|
3.969
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr5_-_111091947
|
3.916
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_183387501
|
3.840
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr4_+_110749149
|
3.800
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr5_+_140566655
|
3.798
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr14_-_58618807
|
3.790
|
NM_001001872
|
C14orf37
|
chromosome 14 open reading frame 37
|
|
chr2_+_237478379
|
3.769
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr1_+_90098643
|
3.768
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr10_-_13749622
|
3.761
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_-_111846192
|
3.737
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr19_-_18717467
|
3.737
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr7_+_29519485
|
3.721
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr6_+_121756744
|
3.714
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr3_+_39509069
|
3.711
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr3_+_141105283
|
3.693
|
|
|
|
|
chr2_-_161056573
|
3.692
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr4_+_78078332
|
3.680
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr8_+_32504250
|
3.680
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr3_-_123339178
|
3.676
|
|
MYLK
|
myosin light chain kinase
|
|
chr6_-_139613114
|
3.628
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr3_-_123339423
|
3.621
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr5_-_64777703
|
3.618
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr7_-_14030864
|
3.592
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chrX_+_105937067
|
3.556
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr14_+_77228132
|
3.533
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr19_+_20011700
|
3.516
|
NM_031218
|
ZNF93
|
zinc finger protein 93
|
|
chr2_+_149633173
|
3.507
|
|
|
|
|
chr15_-_50411405
|
3.503
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr1_+_92495484
|
3.501
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr14_-_92413738
|
3.498
|
|
FBLN5
|
fibulin 5
|
|
chr12_-_42877785
|
3.491
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr3_+_19189969
|
3.464
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr7_-_117513466
|
3.441
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr3_+_181417387
|
3.428
|
|
SOX2-OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr5_-_16936371
|
3.396
|
NM_012334
|
MYO10
|
myosin X
|
|
chr5_+_140165875
|
3.327
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr6_+_13924676
|
3.315
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr20_-_56284945
|
3.296
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr9_-_77502635
|
3.291
|
NM_001177310
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr7_+_26400503
|
3.284
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr5_-_111092865
|
3.256
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_20181869
|
3.252
|
NM_001029865
|
DBX1
|
developing brain homeobox 1
|
|
chr19_-_17375522
|
3.250
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr6_-_47253962
|
3.219
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr5_-_16936290
|
3.217
|
|
MYO10
|
myosin X
|
|
chr18_+_11751470
|
3.184
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr14_+_85996454
|
3.174
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_-_95392449
|
3.128
|
|
CNN3
|
calponin 3, acidic
|
|
chr12_-_111358353
|
3.125
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr9_-_21482311
|
3.125
|
NM_176891
|
IFNE
|
interferon, epsilon
|
|
chr17_-_39675078
|
3.114
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr2_+_166428845
|
3.111
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr7_-_14031049
|
3.086
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr1_+_90098693
|
3.081
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr8_+_75736771
|
3.058
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr16_+_69141442
|
3.048
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr10_+_11207053
|
3.035
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_+_111260925
|
3.028
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr17_+_56065408
|
3.018
|
|
|
|
|
chr5_-_168271852
|
2.978
|
|
|
|
|
chr12_-_42877743
|
2.970
|
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chrX_-_128788913
|
2.943
|
NM_017413
|
APLN
|
apelin
|
|
chr10_-_23633771
|
2.938
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr2_-_110873533
|
2.927
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr1_+_164528914
|
2.925
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr14_+_85996512
|
2.921
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_+_107288199
|
2.919
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr3_-_121740968
|
2.918
|
NM_001199799
NM_001199800
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr3_-_149510609
|
2.915
|
NM_001144960
|
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1
|
|
chr13_+_53602829
|
2.907
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr12_-_113909876
|
2.903
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr2_-_110874142
|
2.900
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr5_-_111092918
|
2.891
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_11779279
|
2.888
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr15_-_42186274
|
2.880
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr8_-_87755880
|
2.864
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr17_-_33759518
|
2.847
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr1_+_203096835
|
2.833
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr11_-_85430117
|
2.833
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_+_2622134
|
2.831
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr22_+_31090792
|
2.823
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr5_-_100238870
|
2.818
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr1_+_90098624
|
2.809
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr4_-_184243578
|
2.798
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr2_-_208031570
|
2.784
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr11_+_130184326
|
2.776
|
|
FLJ34521
|
uncharacterized LOC646383
|
|
chr11_+_111808017
|
2.757
|
|
DIXDC1
|
DIX domain containing 1
|
|
chr11_+_57365026
|
2.750
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr15_+_80696569
|
2.722
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr6_-_32163890
|
2.721
|
|
NOTCH4
|
notch 4
|
|
chr7_+_63773185
|
2.721
|
NM_001170905
|
ZNF736
|
zinc finger protein 736
|
|
chr5_+_140625146
|
2.713
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chr7_+_106505739
|
2.712
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr4_+_76649739
|
2.696
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr8_-_139926235
|
2.691
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr7_+_89843133
|
2.674
|
NM_001040666
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr5_+_140588267
|
2.672
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr9_+_2622099
|
2.669
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr11_-_111782447
|
2.667
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr15_-_82338348
|
2.663
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr5_-_39424930
|
2.660
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|