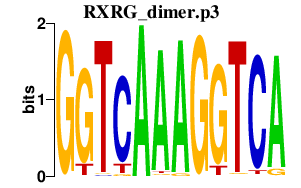
|
chr19_+_45116921
|
7.285
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr19_-_55580896
|
5.521
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr1_-_29309092
|
3.897
|
|
|
|
|
chr5_+_92918924
|
3.223
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr5_-_42811959
|
2.950
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr1_+_78354199
|
2.779
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr1_-_155881063
|
2.628
|
|
RIT1
|
Ras-like without CAAX 1
|
|
chr7_+_97841565
|
2.564
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr19_+_54606141
|
2.555
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr4_+_30721865
|
2.285
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr1_-_155881163
|
2.217
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr3_-_48936227
|
2.086
|
NM_000387
|
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20
|
|
chr19_+_44100734
|
2.017
|
NM_001145347
|
ZNF576
|
zinc finger protein 576
|
|
chr4_+_30722191
|
1.956
|
|
PCDH7
|
protocadherin 7
|
|
chr19_-_45579687
|
1.893
|
NM_145288
|
ZNF296
|
zinc finger protein 296
|
|
chr1_-_151138330
|
1.819
|
NM_212551
NM_001136543
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1
|
|
chr12_-_118810550
|
1.776
|
|
TAOK3
|
TAO kinase 3
|
|
chr11_-_116708334
|
1.750
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr22_-_24059524
|
1.668
|
|
GUSBP11
|
glucuronidase, beta pseudogene 11
|
|
chr1_+_61547625
|
1.645
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr19_+_45394642
|
1.629
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr11_+_118868861
|
1.627
|
|
CCDC84
|
coiled-coil domain containing 84
|
|
chr11_+_118868841
|
1.573
|
NM_198489
|
CCDC84
|
coiled-coil domain containing 84
|
|
chr22_-_24059502
|
1.572
|
|
GUSBP11
|
glucuronidase, beta pseudogene 11
|
|
chr19_+_45394522
|
1.565
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr12_-_118810712
|
1.546
|
NM_016281
|
TAOK3
|
TAO kinase 3
|
|
chr11_-_61658853
|
1.506
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr19_+_39616384
|
1.456
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr17_+_7812703
|
1.429
|
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr9_+_88897112
|
1.427
|
|
|
|
|
chr3_-_113464640
|
1.425
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr19_+_6372720
|
1.424
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr7_+_143079354
|
1.407
|
|
ZYX
|
zyxin
|
|
chr11_+_116700620
|
1.406
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr7_-_139763520
|
1.394
|
NM_022750
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr19_+_6372443
|
1.370
|
NM_032306
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr19_-_38806381
|
1.370
|
NM_001039671
NM_001145461
NM_001145462
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr1_-_43855473
|
1.342
|
NM_052877
NM_201542
|
MED8
|
mediator complex subunit 8
|
|
chr19_+_39390567
|
1.337
|
NM_002503
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr2_-_64246431
|
1.325
|
|
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae)
|
|
chr19_+_6372805
|
1.311
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr1_-_43855448
|
1.307
|
|
MED8
|
mediator complex subunit 8
|
|
chr6_+_143999058
|
1.265
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr22_-_24059436
|
1.252
|
|
|
|
|
chr12_+_96252705
|
1.242
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr7_+_143079925
|
1.213
|
|
ZYX
|
zyxin
|
|
chr18_+_54318561
|
1.207
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr1_+_12079552
|
1.204
|
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr22_-_41864656
|
1.177
|
NM_032758
|
PHF5A
|
PHD finger protein 5A
|
|
chr13_+_100741268
|
1.158
|
NM_000282
NM_001127692
NM_001178004
|
PCCA
|
propionyl CoA carboxylase, alpha polypeptide
|
|
chr2_-_176046120
|
1.149
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr2_+_178257371
|
1.133
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr19_+_10362912
|
1.127
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr17_-_8198605
|
1.114
|
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr2_+_178257488
|
1.111
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr19_+_39390618
|
1.092
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr2_-_28113211
|
1.076
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr6_+_143771779
|
1.060
|
NM_003630
|
PEX3
|
peroxisomal biogenesis factor 3
|
|
chr7_+_129015483
|
1.059
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_45939361
|
1.051
|
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr12_+_96252770
|
1.018
|
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr2_-_24307179
|
0.998
|
NM_001206802
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr19_+_10362886
|
0.992
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chrX_-_122866881
|
0.976
|
|
THOC2
|
THO complex 2
|
|
chr3_-_113465101
|
0.963
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr21_+_45432202
|
0.953
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chrX_-_122866888
|
0.933
|
NM_001081550
|
THOC2
|
THO complex 2
|
|
chr1_+_12079522
|
0.928
|
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr6_-_146285225
|
0.922
|
NM_001042683
NM_173082
|
SHPRH
|
SNF2 histone linker PHD RING helicase
|
|
chr1_-_120484230
|
0.894
|
|
NOTCH2
|
notch 2
|
|
chr17_-_8198124
|
0.888
|
NM_201520
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr16_-_30596808
|
0.885
|
NM_152458
|
ZNF785
|
zinc finger protein 785
|
|
chr4_-_7069755
|
0.881
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr8_+_22423170
|
0.876
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr14_-_95623606
|
0.874
|
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_+_120125141
|
0.872
|
NM_001079863
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein)
|
|
chr20_-_32899607
|
0.869
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr4_-_4543690
|
0.869
|
|
STX18
|
syntaxin 18
|
|
chr19_+_39390570
|
0.858
|
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr19_+_2328628
|
0.846
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr16_-_47007567
|
0.837
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr15_+_96873845
|
0.836
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr11_-_45939568
|
0.797
|
NM_004813
NM_057174
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr19_+_16308664
|
0.796
|
NM_001130524
NM_032493
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr5_+_1801493
|
0.789
|
NM_004553
|
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase)
|
|
chr16_-_4401265
|
0.783
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr17_+_41150465
|
0.780
|
|
RPL27
|
ribosomal protein L27
|
|
chr2_+_73441344
|
0.775
|
NM_006062
|
SMYD5
|
SMYD family member 5
|
|
chr2_-_207024103
|
0.768
|
NM_001199981
NM_001199982
NM_001199983
NM_005006
|
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase)
|
|
chr20_+_36662138
|
0.767
|
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr17_+_41150409
|
0.764
|
NM_000988
|
RPL27
|
ribosomal protein L27
|
|
chr15_+_43885251
|
0.763
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr4_-_4543719
|
0.755
|
|
STX18
|
syntaxin 18
|
|
chr20_+_36662207
|
0.753
|
|
|
|
|
chr3_-_113464991
|
0.725
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr12_-_51422012
|
0.724
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr22_+_30163351
|
0.721
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr16_-_47007483
|
0.720
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr7_-_74489636
|
0.716
|
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr19_-_38806546
|
0.707
|
NM_001039672
NM_001039673
NM_001145463
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr10_-_96829074
|
0.706
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr2_+_220299699
|
0.690
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr20_-_44485953
|
0.685
|
NM_005469
|
ACOT8
|
acyl-CoA thioesterase 8
|
|
chr8_-_80942488
|
0.670
|
NM_014018
|
MRPS28
|
mitochondrial ribosomal protein S28
|
|
chr16_+_4714709
|
0.666
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr19_+_45394459
|
0.655
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr7_-_72936447
|
0.653
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr1_-_101491308
|
0.643
|
NM_001077394
NM_001077395
NM_015958
|
DPH5
|
DPH5 homolog (S. cerevisiae)
|
|
chr14_-_95623665
|
0.629
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_-_194392913
|
0.617
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chrX_-_108868250
|
0.616
|
NM_012282
|
KCNE1L
|
KCNE1-like
|
|
chr1_+_151138497
|
0.601
|
NM_001204856
NM_024041
|
SCNM1
|
sodium channel modifier 1
|
|
chr6_-_146285170
|
0.593
|
|
SHPRH
|
SNF2 histone linker PHD RING helicase
|
|
chr12_-_95467104
|
0.591
|
NM_001032287
NM_001127362
NM_003297
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1
|
|
chr2_-_220034706
|
0.580
|
NM_001144889
NM_001144890
NM_144712
|
SLC23A3
|
solute carrier family 23 (nucleobase transporters), member 3
|
|
chr7_-_74489686
|
0.580
|
NM_030798
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr20_+_44509847
|
0.580
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr17_+_7123246
|
0.561
|
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr12_+_67663364
|
0.561
|
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr8_-_82607206
|
0.553
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr7_-_74489572
|
0.551
|
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr12_+_121416294
|
0.542
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr1_+_43855549
|
0.540
|
NM_015284
|
SZT2
|
seizure threshold 2 homolog (mouse)
|
|
chr1_-_6761878
|
0.540
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr1_-_6761839
|
0.539
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr7_-_72936538
|
0.532
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr2_+_207024317
|
0.520
|
NM_001037663
NM_001959
NM_021121
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr5_+_132209354
|
0.520
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr1_-_6420719
|
0.517
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr4_+_4543718
|
0.512
|
|
LOC100507266
|
uncharacterized LOC100507266
|
|
chr6_-_146285545
|
0.505
|
|
SHPRH
|
SNF2 histone linker PHD RING helicase
|
|
chr19_+_10197023
|
0.502
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chrX_-_18372777
|
0.501
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr10_+_120967094
|
0.497
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr3_-_150264134
|
0.492
|
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr3_-_194392938
|
0.490
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr22_+_41865112
|
0.480
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr4_-_7069801
|
0.476
|
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr6_+_30594607
|
0.474
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr7_-_72936592
|
0.466
|
NM_032408
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr17_-_7123368
|
0.465
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr2_-_191399449
|
0.464
|
NM_001142645
|
TMEM194B
|
transmembrane protein 194B
|
|
chr19_+_45394569
|
0.456
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr2_+_159313407
|
0.454
|
|
PKP4
|
plakophilin 4
|
|
chr17_-_1395950
|
0.453
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr3_-_150264234
|
0.448
|
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr6_-_31138180
|
0.447
|
NM_002701
|
POU5F1
POU5F1B
POU5F1P3
POU5F1P4
|
POU class 5 homeobox 1
POU class 5 homeobox 1B
POU class 5 homeobox 1 pseudogene 3
POU class 5 homeobox 1 pseudogene 4
|
|
chr6_+_31623688
|
0.447
|
|
APOM
|
apolipoprotein M
|
|
chr2_+_207024614
|
0.436
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr22_-_44893179
|
0.433
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr19_+_10196972
|
0.432
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr1_+_43855599
|
0.427
|
|
SZT2
|
seizure threshold 2 homolog (mouse)
|
|
chr8_+_22436641
|
0.424
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr1_-_55089068
|
0.421
|
NM_176782
|
FAM151A
|
family with sequence similarity 151, member A
|
|
chr3_+_37284680
|
0.419
|
NM_001172713
NM_002078
|
GOLGA4
|
golgin A4
|
|
chr1_-_95538443
|
0.417
|
NM_144988
|
ALG14
|
asparagine-linked glycosylation 14 homolog (S. cerevisiae)
|
|
chr17_-_2614926
|
0.415
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chrX_+_100354797
|
0.412
|
NM_006733
|
CENPI
|
centromere protein I
|
|
chr3_-_14166370
|
0.411
|
NM_001098502
NM_144636
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr2_+_28113481
|
0.411
|
NM_199193
NM_199194
NM_004899
NM_199191
NM_199192
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr3_-_14166292
|
0.410
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr3_-_150264264
|
0.408
|
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr4_+_156680603
|
0.402
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr17_+_38279468
|
0.400
|
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr20_+_36661849
|
0.400
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr19_+_36239261
|
0.397
|
NM_019104
|
LIN37
|
lin-37 homolog (C. elegans)
|
|
chr12_-_7079616
|
0.396
|
|
PHB2
|
prohibitin 2
|
|
chr2_+_109065576
|
0.392
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr5_+_118965253
|
0.388
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr12_+_67663147
|
0.387
|
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr2_+_109065665
|
0.386
|
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr1_-_6761897
|
0.383
|
NM_018198
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr6_+_142468382
|
0.365
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr19_-_1020995
|
0.365
|
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr1_-_226374401
|
0.364
|
NM_022735
|
ACBD3
|
acyl-CoA binding domain containing 3
|
|
chr5_+_14664803
|
0.361
|
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr6_-_2840683
|
0.361
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr9_-_139137646
|
0.360
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr2_-_178257380
|
0.355
|
|
LOC100130691
|
uncharacterized LOC100130691
|
|
chr19_+_2328679
|
0.354
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr1_-_40042461
|
0.353
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr2_-_191399415
|
0.347
|
|
TMEM194B
|
transmembrane protein 194B
|
|
chr12_+_132413776
|
0.342
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr18_-_51750460
|
0.340
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr2_-_198364997
|
0.337
|
NM_199440
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr2_+_28113598
|
0.336
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr1_-_40042519
|
0.336
|
NM_001135653
NM_001135654
NM_003819
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr10_+_114133838
|
0.334
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_-_39390168
|
0.329
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr16_+_81272293
|
0.327
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr2_-_198364522
|
0.324
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr6_+_142468401
|
0.324
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr5_+_14664764
|
0.321
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr6_+_31623668
|
0.316
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr2_+_28113612
|
0.314
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr4_-_102268333
|
0.312
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr7_-_25164902
|
0.310
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr3_-_49314281
|
0.308
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr12_+_50135953
|
0.306
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr19_-_2328566
|
0.304
|
NM_016199
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|