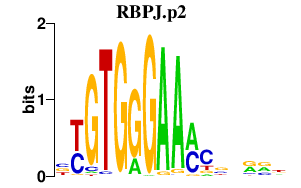
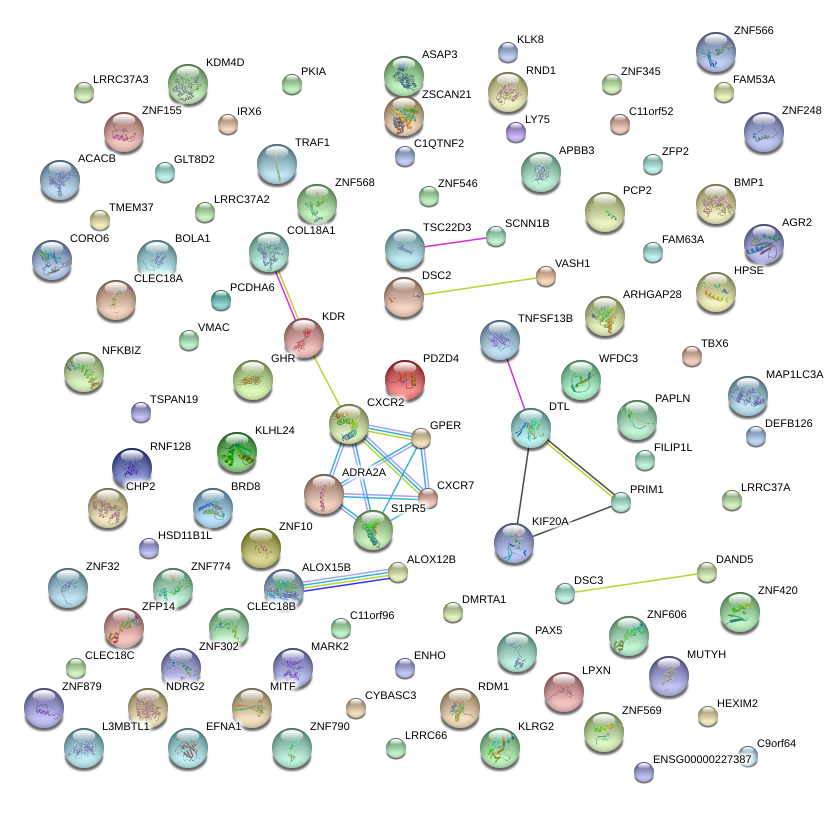
|
chr10_-_44144151
|
11.125
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr10_-_44144266
|
8.119
|
NM_006973
|
ZNF32
|
zinc finger protein 32
|
|
chr19_-_51504799
|
5.369
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr2_-_160761192
|
5.255
|
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr19_+_44488331
|
4.939
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chr11_+_43963809
|
4.420
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr19_+_37407230
|
3.769
|
NM_001204835
NM_001204836
NM_001204837
NM_001204838
NM_001204839
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr5_-_137514299
|
3.759
|
NM_001164326
NM_006696
NM_139199
|
BRD8
|
bromodomain containing 8
|
|
chr2_-_160761266
|
3.711
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr17_+_7942351
|
3.465
|
NM_001039130
NM_001039131
NM_001141
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B
|
|
chr5_+_137514753
|
3.443
|
|
KIF20A
|
kinesin family member 20A
|
|
chr7_-_139168408
|
3.152
|
NM_198508
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2
|
|
chr17_-_34257730
|
3.103
|
NM_001034836
NM_001163120
NM_001163121
NM_145654
|
RDM1
|
RAD52 motif 1
|
|
chr19_+_36980524
|
2.983
|
|
LOC728752
|
uncharacterized LOC728752
|
|
chr4_-_55991746
|
2.926
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr20_-_1165116
|
2.834
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr9_-_123688352
|
2.734
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr19_+_37341259
|
2.705
|
NM_003419
|
ZNF345
|
zinc finger protein 345
|
|
chrX_-_153095996
|
2.672
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr19_+_35168558
|
2.634
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr17_-_27949814
|
2.507
|
|
CORO6
|
coronin 6
|
|
chr17_-_34257285
|
2.483
|
NM_001163122
NM_001163124
NM_001163125
NM_001163130
|
RDM1
|
RAD52 motif 1
|
|
chr1_+_149871154
|
2.435
|
NM_016074
|
BOLA1
|
bolA homolog 1 (E. coli)
|
|
chr5_+_178450775
|
2.376
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr19_-_37407079
|
2.364
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr19_-_37406918
|
2.273
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr16_+_23765947
|
2.267
|
NM_022097
|
CHP2
|
calcineurin B homologous protein 2
|
|
chr2_+_237478379
|
2.263
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr5_+_42423811
|
2.158
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr12_-_104443698
|
2.131
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr9_-_34522972
|
2.088
|
NM_198573
|
ENHO
|
energy homeostasis associated
|
|
chr5_+_180257979
|
2.054
|
|
LOC729678
|
uncharacterized LOC729678
|
|
chr19_-_58514136
|
1.995
|
|
ZNF606
|
zinc finger protein 606
|
|
chr11_+_111789600
|
1.955
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr14_+_77228940
|
1.954
|
|
VASH1
|
vasohibin 1
|
|
chr19_-_37341168
|
1.919
|
NM_001242801
|
ZNF790
|
zinc finger protein 790
|
|
chr1_-_45805606
|
1.873
|
NM_001048174
NM_001048172
NM_001048173
NM_001048171
NM_001128425
NM_012222
|
MUTYH
|
mutY homolog (E. coli)
|
|
chr16_-_30103204
|
1.853
|
NM_004608
|
TBX6
|
T-box 6
|
|
chr19_-_37958338
|
1.844
|
NM_152484
|
ZNF569
|
zinc finger protein 569
|
|
chr5_+_180257936
|
1.823
|
|
|
|
|
chr19_-_10625679
|
1.779
|
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr14_-_21493122
|
1.761
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr19_+_37569307
|
1.760
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|
|
chr12_+_133707212
|
1.730
|
NM_015394
|
ZNF10
|
zinc finger protein 10
|
|
chr10_-_38146221
|
1.705
|
NM_021045
|
ZNF248
|
zinc finger protein 248
|
|
chr12_+_109577201
|
1.686
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr19_+_5681016
|
1.684
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr17_-_7991020
|
1.629
|
NM_001139
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type
|
|
chrX_+_105969875
|
1.623
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr5_+_137514416
|
1.621
|
NM_005733
|
KIF20A
|
kinesin family member 20A
|
|
chr9_-_123689172
|
1.600
|
NM_005658
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr18_+_6834431
|
1.594
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr19_+_40502942
|
1.577
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chr20_+_42143045
|
1.551
|
NM_015478
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila)
|
|
chr19_-_36980336
|
1.534
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr11_+_94706780
|
1.529
|
NM_018039
|
KDM4D
|
lysine (K)-specific demethylase 4D
|
|
chr20_-_44420538
|
1.498
|
NM_080614
|
WFDC3
|
WAP four-disulfide core domain 3
|
|
chr14_+_73704204
|
1.493
|
NM_173462
|
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein
|
|
chr12_-_49259549
|
1.493
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr9_-_123676849
|
1.483
|
NM_001190947
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr19_+_5681139
|
1.483
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chrX_-_153095332
|
1.474
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr8_+_79428335
|
1.461
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr9_-_86571652
|
1.458
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr3_-_99833348
|
1.431
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr2_+_218990735
|
1.417
|
NM_001557
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr5_+_178322915
|
1.407
|
NM_030613
|
ZFP2
|
zinc finger protein 2 homolog (mouse)
|
|
chrX_-_153095841
|
1.403
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr5_+_140306301
|
1.399
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr20_+_123230
|
1.361
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr6_+_43968303
|
1.343
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr5_-_139943829
|
1.293
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr1_-_150980840
|
1.290
|
NM_001163258
NM_018379
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr7_-_16844606
|
1.287
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr12_-_104443914
|
1.284
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr12_-_57146081
|
1.256
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr4_-_84256304
|
1.236
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr20_+_33146500
|
1.228
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr21_+_46875402
|
1.224
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr11_-_61129462
|
1.218
|
NM_001161454
NM_153611
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr3_+_101546833
|
1.217
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr17_+_43238263
|
1.216
|
NM_144608
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2
|
|
chr4_-_52883785
|
1.215
|
NM_001024611
|
LRRC66
|
leucine rich repeat containing 66
|
|
chr19_-_10628606
|
1.200
|
NM_001166215
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr19_-_36870064
|
1.189
|
NM_020917
|
ZFP14
|
zinc finger protein 14 homolog (mouse)
|
|
chr19_-_7698569
|
1.165
|
NM_174895
|
PCP2
|
Purkinje cell protein 2
|
|
chr11_-_58345593
|
1.165
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr19_+_5904842
|
1.163
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr15_+_90895476
|
1.152
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr4_-_84256012
|
1.138
|
NM_001098540
|
HPSE
|
heparanase
|
|
chr9_+_22446783
|
1.129
|
NM_022160
|
DMRTA1
|
DMRT-like family A1
|
|
chr3_+_183353355
|
1.120
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr9_-_86571631
|
1.101
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr16_+_69984809
|
1.100
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr1_+_155100304
|
1.099
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr2_+_120189342
|
1.095
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr17_+_44590075
|
1.087
|
NM_001006607
|
LRRC37A2
LRRC37A
|
leucine rich repeat containing 37, member A2
leucine rich repeat containing 37A
|
|
chr16_+_55358470
|
1.085
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr18_-_28622624
|
1.083
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr12_-_85430054
|
1.066
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr16_+_23313590
|
1.059
|
NM_000336
|
SCNN1B
|
sodium channel, nonvoltage-gated 1, beta
|
|
chr1_-_23810627
|
1.044
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr7_+_1127722
|
1.042
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr19_-_36980758
|
1.027
|
|
ZNF566
|
zinc finger protein 566
|
|
chr5_-_159797618
|
1.011
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chrX_-_106960268
|
1.005
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_+_69812961
|
1.004
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_212208918
|
0.982
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr11_-_61129273
|
0.977
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr4_-_1685717
|
0.974
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr19_+_13080431
|
0.968
|
NM_152654
|
DAND5
|
DAN domain family, member 5
|
|
chr12_+_70759974
|
0.963
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr1_+_142618798
|
0.958
|
|
|
|
|
chr20_-_62130390
|
0.956
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr16_+_23690191
|
0.955
|
NM_005030
|
PLK1
|
polo-like kinase 1
|
|
chr5_+_140792507
|
0.953
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr1_+_36689996
|
0.951
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_-_51456159
|
0.951
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr16_+_23690234
|
0.946
|
|
PLK1
|
polo-like kinase 1
|
|
chr22_-_31688329
|
0.938
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr10_+_94352809
|
0.936
|
NM_004523
|
KIF11
|
kinesin family member 11
|
|
chr17_+_80693398
|
0.935
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr3_+_69812620
|
0.935
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr16_+_23690212
|
0.932
|
|
PLK1
|
polo-like kinase 1
|
|
chr11_-_61129352
|
0.931
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr20_+_8112762
|
0.929
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr17_-_42092319
|
0.927
|
NM_032376
|
TMEM101
|
transmembrane protein 101
|
|
chr19_+_50706868
|
0.924
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr19_-_44405934
|
0.923
|
|
LOC100505715
|
uncharacterized LOC100505715
|
|
chr1_-_93811322
|
0.909
|
|
LOC100131564
|
uncharacterized LOC100131564
|
|
chr16_+_23690190
|
0.906
|
|
PLK1
|
polo-like kinase 1
|
|
chr1_+_212209112
|
0.905
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr16_+_23690160
|
0.905
|
|
PLK1
|
polo-like kinase 1
|
|
chr17_+_1648262
|
0.901
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr4_-_84255895
|
0.895
|
NM_001199830
|
HPSE
|
heparanase
|
|
chr16_+_640172
|
0.894
|
NM_021168
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr19_+_14544100
|
0.885
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr17_+_80317122
|
0.884
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr4_-_84255999
|
0.880
|
|
HPSE
|
heparanase
|
|
chr1_+_212209129
|
0.877
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr10_+_111765725
|
0.876
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr14_-_54421269
|
0.871
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr20_-_33460558
|
0.864
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr11_-_113746205
|
0.860
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr9_+_90112754
|
0.852
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr9_-_86432546
|
0.850
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr1_+_201979689
|
0.846
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr9_-_85677784
|
0.841
|
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr1_+_152758752
|
0.839
|
NM_178353
|
LCE1E
|
late cornified envelope 1E
|
|
chr15_+_67835002
|
0.837
|
NM_002757
NM_145160
|
MAP2K5
|
mitogen-activated protein kinase kinase 5
|
|
chr9_+_136325086
|
0.832
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr7_-_99381703
|
0.832
|
NM_001202855
NM_017460
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4
|
|
chr3_+_112931374
|
0.829
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr7_+_8008416
|
0.828
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr17_-_46035109
|
0.820
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr21_-_43187197
|
0.818
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr19_-_58892382
|
0.817
|
NM_001129730
NM_138466
|
ZNF837
|
zinc finger protein 837
|
|
chr4_-_57976516
|
0.814
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr16_+_639356
|
0.814
|
NM_001172663
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr3_-_187452669
|
0.814
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chrX_+_123094377
|
0.811
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr2_+_48844936
|
0.809
|
NM_001193487
NM_006872
|
GTF2A1L
|
general transcription factor IIA, 1-like
|
|
chr22_+_27068690
|
0.802
|
|
LOC100507637
|
uncharacterized LOC100507637
|
|
chr6_+_79787548
|
0.796
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr5_-_112630557
|
0.793
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr15_+_45028559
|
0.791
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr6_-_36355342
|
0.789
|
NM_001207038
NM_001207035
NM_001207036
NM_001207037
NM_001207039
NM_001207040
NM_001207041
NM_016135
|
ETV7
|
ets variant 7
|
|
chrX_+_24483343
|
0.788
|
NM_001142386
NM_005391
|
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3
|
|
chr1_-_186344241
|
0.787
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr15_+_40697977
|
0.786
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr11_-_82782883
|
0.784
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr17_+_46125675
|
0.776
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr11_-_82782870
|
0.771
|
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr15_+_43622871
|
0.770
|
NM_001012969
NM_001159280
|
ADAL
|
adenosine deaminase-like
|
|
chr10_+_94352873
|
0.768
|
|
KIF11
|
kinesin family member 11
|
|
chr10_-_72141347
|
0.768
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr17_+_39994042
|
0.764
|
NM_152467
|
KLHL10
|
kelch-like 10 (Drosophila)
|
|
chr12_-_59314069
|
0.764
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr17_-_26926044
|
0.761
|
NM_006461
|
SPAG5
|
sperm associated antigen 5
|
|
chr19_-_58514713
|
0.759
|
NM_025027
|
ZNF606
|
zinc finger protein 606
|
|
chr15_+_40698000
|
0.758
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr1_-_182361311
|
0.754
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chrX_+_106045880
|
0.754
|
NM_017752
NM_198881
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chr15_+_40698018
|
0.752
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr1_+_36690038
|
0.748
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr17_-_42264075
|
0.746
|
NM_178542
|
C17orf65
|
chromosome 17 open reading frame 65
|
|
chr3_-_123339423
|
0.743
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr12_+_109551041
|
0.740
|
|
|
|
|
chr6_+_52226866
|
0.740
|
NM_133367
|
PAQR8
|
progestin and adipoQ receptor family member VIII
|
|
chr16_+_2039945
|
0.739
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chrX_+_134555867
|
0.733
|
|
LINC00086
|
long intergenic non-protein coding RNA 86
|
|
chr5_+_76382458
|
0.730
|
|
LOC728723
|
uncharacterized LOC728723
|
|
chr10_+_75673346
|
0.729
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr18_+_61442611
|
0.726
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr9_-_85678042
|
0.722
|
NM_152573
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr20_+_44637539
|
0.722
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr7_-_150497398
|
0.720
|
NM_001101311
NM_001101312
NM_001101314
|
TMEM176B
|
transmembrane protein 176B
|
|
chr15_+_32322690
|
0.718
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chrX_-_47930482
|
0.717
|
NM_001037735
|
ZNF630
|
zinc finger protein 630
|
|
chr7_+_79764087
|
0.715
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|