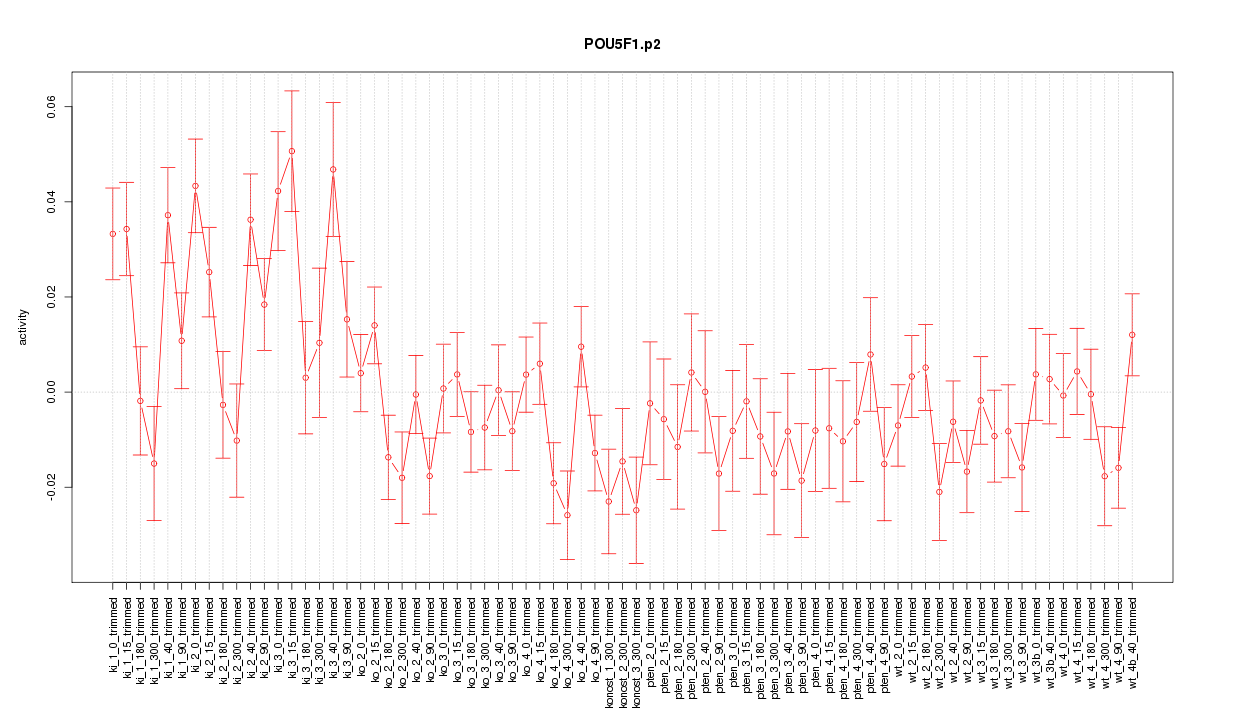
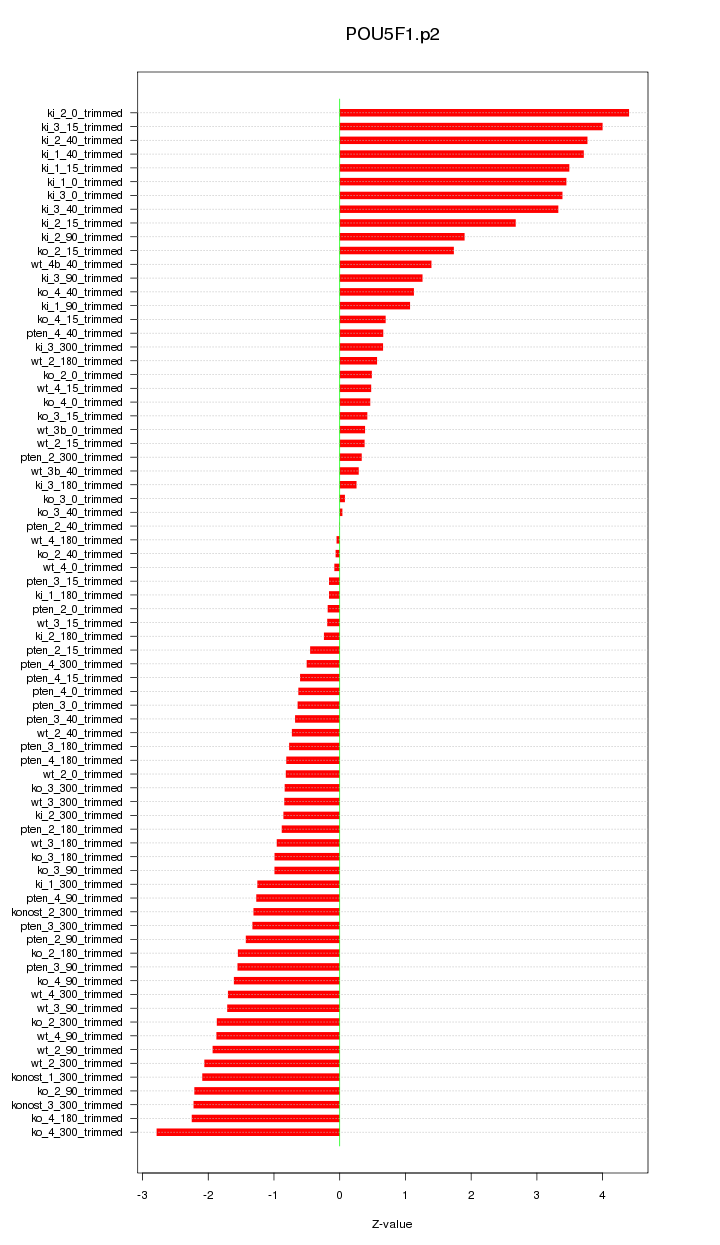
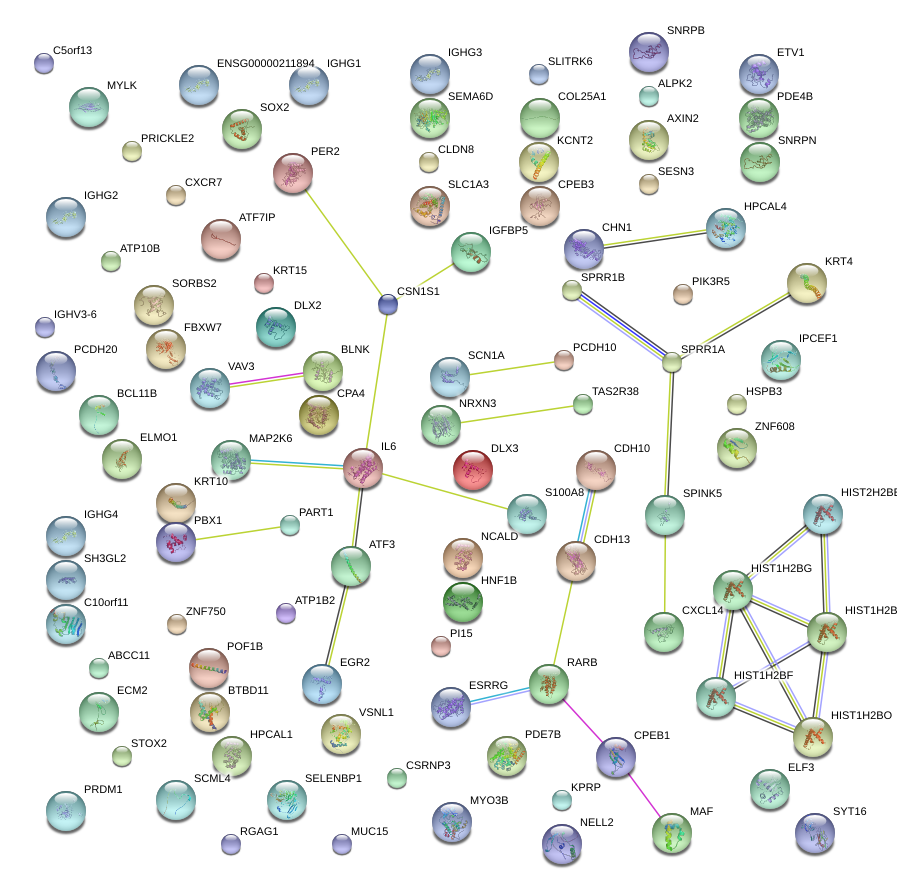
|
chr17_+_67410825
|
36.184
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr16_-_79634610
|
17.379
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr17_-_38978824
|
15.171
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr1_+_164528914
|
13.330
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_-_108231122
|
12.248
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr10_-_64576123
|
11.618
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64576074
|
11.375
|
|
EGR2
|
early growth response 2
|
|
chr10_-_64578926
|
10.429
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr1_+_61542790
|
10.298
|
|
|
|
|
chr1_-_153363544
|
9.734
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr10_-_98031154
|
8.631
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr3_-_64211111
|
7.738
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_164528586
|
7.691
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_-_26593667
|
7.430
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr12_-_45269340
|
7.140
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr17_-_80797889
|
6.506
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr15_+_25101697
|
6.469
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr9_+_17578952
|
6.407
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr1_+_201979689
|
6.403
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_+_79745653
|
6.388
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr1_+_201979747
|
6.037
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr2_-_175712276
|
5.790
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr1_-_108507474
|
5.764
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr17_-_48072566
|
5.685
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr14_-_99737821
|
5.527
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr17_-_39675078
|
5.471
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr2_+_237478379
|
5.417
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr5_-_124080773
|
5.249
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr16_+_82660390
|
5.173
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr1_+_152730505
|
5.146
|
NM_001025231
|
KPRP
|
keratinocyte proline-rich protein
|
|
chr2_+_171034654
|
5.107
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr1_+_212782103
|
5.105
|
|
ATF3
|
activating transcription factor 3
|
|
chr11_-_94964209
|
5.062
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr4_-_153303663
|
4.961
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr5_-_160279045
|
4.949
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr1_+_66458389
|
4.922
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_-_217263195
|
4.892
|
NM_001243505
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_-_123603144
|
4.749
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr8_+_75736771
|
4.681
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr18_-_56296188
|
4.659
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr5_+_53751430
|
4.575
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr6_-_26216871
|
4.566
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr15_+_25068793
|
4.479
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr17_-_63557615
|
4.457
|
NM_004655
|
AXIN2
|
axin 2
|
|
chr16_+_82660641
|
4.405
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr7_-_141673572
|
4.372
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr17_+_7554415
|
4.367
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr16_+_82660573
|
4.334
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chrX_-_84634742
|
4.328
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr5_+_59784004
|
4.242
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr5_-_134914968
|
4.235
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr7_-_14029286
|
4.235
|
|
ETV1
|
ets variant 1
|
|
chr1_-_151345156
|
4.229
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr22_+_22676814
|
4.225
|
|
|
|
|
chrX_-_84634664
|
4.177
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr6_+_136172802
|
4.144
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_+_184826427
|
4.048
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr5_-_111091947
|
4.041
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_-_186733372
|
4.004
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_+_17721983
|
3.903
|
|
VSNL1
|
visinin-like 1
|
|
chr5_-_111092918
|
3.864
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_53207843
|
3.847
|
NM_002272
|
KRT4
|
keratin 4
|
|
chr2_+_166326156
|
3.847
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr7_-_37024664
|
3.841
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr2_+_17721806
|
3.804
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr6_+_135516879
|
3.793
|
|
|
|
|
chr10_-_94003002
|
3.773
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr14_+_62462540
|
3.762
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr2_-_167005641
|
3.762
|
NM_001202435
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr1_-_196577438
|
3.752
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr11_+_65271145
|
3.750
|
|
|
|
|
chr13_-_61989209
|
3.742
|
|
PCDH20
|
protocadherin 20
|
|
chr15_+_48010685
|
3.722
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_-_111092865
|
3.706
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_129932973
|
3.700
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr2_-_239197091
|
3.660
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr13_+_97999289
|
3.646
|
|
|
|
|
chr5_-_24645084
|
3.631
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr1_-_196577676
|
3.603
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr3_+_181430075
|
3.549
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr10_+_77542518
|
3.537
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr5_+_36608430
|
3.506
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_217262946
|
3.488
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_106546736
|
3.483
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr12_+_14518608
|
3.458
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr11_-_56310732
|
3.437
|
NM_001005245
|
OR5M11
|
olfactory receptor, family 5, subfamily M, member 11
|
|
chr17_-_36105005
|
3.409
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr5_-_134914447
|
3.399
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr3_+_25470067
|
3.382
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr14_-_106733392
|
3.363
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr5_-_111312522
|
3.361
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_147443534
|
3.357
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr6_-_108145507
|
3.339
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr21_-_31588242
|
3.319
|
NM_199328
|
CLDN8
|
claudin 8
|
|
chr1_+_153003677
|
3.309
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr3_+_181429711
|
3.305
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr9_-_95298251
|
3.302
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr1_+_212781969
|
3.290
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr17_-_8815799
|
3.279
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr12_+_107712151
|
3.252
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr17_-_36104874
|
3.249
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr7_+_22766765
|
3.247
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr15_-_83240499
|
3.201
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr13_-_86373328
|
3.196
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr4_+_70796798
|
3.190
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chrX_+_109662284
|
3.173
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr1_+_116654375
|
3.139
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr2_-_217540722
|
3.133
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr4_-_110223798
|
3.130
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr6_-_154651214
|
3.129
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr16_-_48269087
|
3.099
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr4_+_134070443
|
3.092
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr1_-_20306908
|
3.087
|
NM_000300
NM_001161727
NM_001161728
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr19_-_18314731
|
3.078
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr12_-_81763127
|
3.074
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_-_153348057
|
3.039
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr2_-_228582698
|
3.038
|
NM_025243
|
SLC19A3
|
solute carrier family 19, member 3
|
|
chr17_+_7554239
|
2.976
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr1_-_177133950
|
2.935
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr19_-_51587466
|
2.926
|
NM_022046
|
KLK14
|
kallikrein-related peptidase 14
|
|
chr13_-_103054027
|
2.901
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr4_-_153273683
|
2.893
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_-_126883568
|
2.883
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr6_-_134497069
|
2.869
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_17575592
|
2.855
|
NM_016233
|
PADI3
|
peptidyl arginine deiminase, type III
|
|
chr11_-_3663484
|
2.850
|
NM_053017
NM_001079536
|
ART5
|
ADP-ribosyltransferase 5
|
|
chr10_+_11206928
|
2.850
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_33722158
|
2.837
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr9_-_75567915
|
2.828
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr12_-_122240827
|
2.827
|
|
|
|
|
chr14_-_99737554
|
2.825
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr12_+_109551041
|
2.815
|
|
|
|
|
chr20_+_56136136
|
2.789
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr22_-_24096551
|
2.786
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr6_+_135517051
|
2.782
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr2_+_189839113
|
2.775
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_-_124990706
|
2.746
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr6_-_26043884
|
2.728
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr7_+_48211054
|
2.726
|
NM_152701
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13
|
|
chr6_-_134498973
|
2.707
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_+_65007418
|
2.696
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr12_-_123187842
|
2.694
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chrX_+_9431314
|
2.690
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_+_248128534
|
2.687
|
NM_001004491
|
OR2AK2
|
olfactory receptor, family 2, subfamily AK, member 2
|
|
chr20_+_17207680
|
2.658
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr20_-_7921068
|
2.654
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr7_+_143140545
|
2.650
|
NM_177437
|
TAS2R60
|
taste receptor, type 2, member 60
|
|
chrX_-_80457440
|
2.645
|
NM_030763
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr15_-_72612497
|
2.616
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr2_+_233734993
|
2.614
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr3_+_48264836
|
2.596
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr6_+_26273143
|
2.592
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr10_-_61122219
|
2.587
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr3_+_46395224
|
2.584
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr14_-_106877689
|
2.551
|
|
IGHV4-31
IGH@
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr6_+_47624222
|
2.547
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr6_-_26124130
|
2.543
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr22_-_45405780
|
2.526
|
NM_001135862
|
PHF21B
|
PHD finger protein 21B
|
|
chr15_-_72612469
|
2.522
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr20_+_306205
|
2.521
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr3_+_134514044
|
2.520
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr4_+_158141735
|
2.513
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr7_-_14029641
|
2.495
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr3_-_193272587
|
2.492
|
NM_032279
|
ATP13A4
|
ATPase type 13A4
|
|
chr8_-_93111915
|
2.485
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_27183225
|
2.480
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr7_+_107110501
|
2.462
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr12_+_93096618
|
2.455
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr3_-_99833348
|
2.447
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr20_+_17207669
|
2.430
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_+_134514300
|
2.421
|
|
EPHB1
|
EPH receptor B1
|
|
chr10_+_11207053
|
2.407
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr7_+_141463896
|
2.402
|
NM_016943
|
TAS2R3
|
taste receptor, type 2, member 3
|
|
chr3_+_127641901
|
2.388
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr18_+_31158540
|
2.381
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr7_-_82791799
|
2.377
|
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr17_-_46655544
|
2.348
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr5_-_146257633
|
2.307
|
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr11_-_115375065
|
2.306
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr2_+_189839132
|
2.306
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_57328044
|
2.305
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr9_-_124990949
|
2.301
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr11_-_118047336
|
2.298
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr7_+_80267972
|
2.291
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_-_177134042
|
2.290
|
|
ASTN1
|
astrotactin 1
|
|
chr6_-_149806025
|
2.288
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr7_-_76829109
|
2.281
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chrX_-_24665454
|
2.277
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr17_+_41052813
|
2.275
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr19_+_14693895
|
2.270
|
NM_001204118
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr4_+_41258941
|
2.263
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr5_-_88179278
|
2.261
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_-_193155723
|
2.253
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr17_-_37381920
|
2.253
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr18_+_61254533
|
2.251
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr3_+_188889762
|
2.248
|
NM_198485
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr2_-_166060552
|
2.244
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr5_-_95748688
|
2.238
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr12_-_45269953
|
2.230
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr15_-_72612127
|
2.227
|
|
CELF6
|
CUGBP, Elav-like family member 6
|