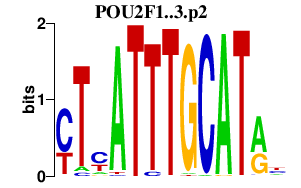
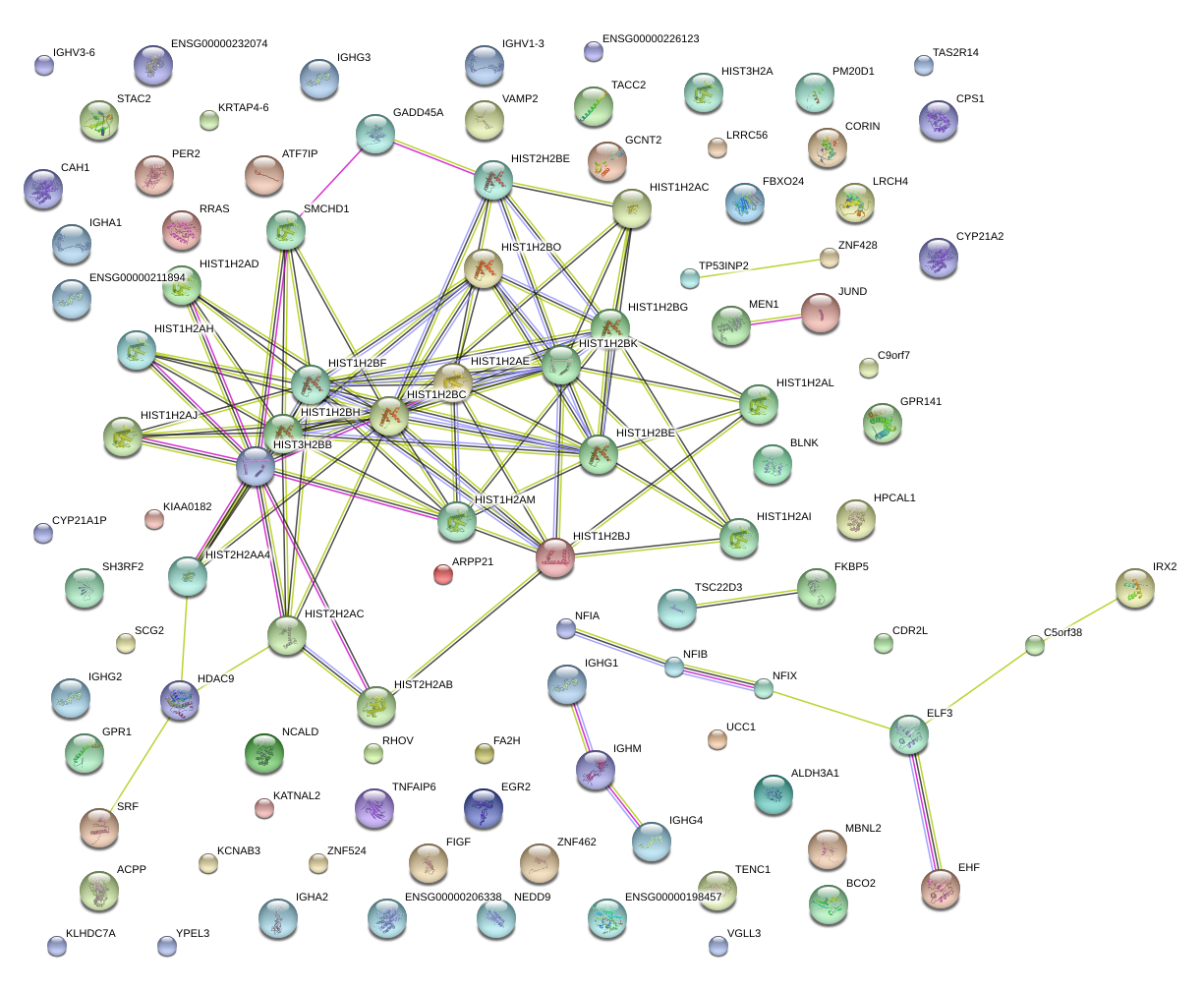
|
chr1_+_201979689
|
31.024
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_-_107211131
|
29.377
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_201979747
|
28.674
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_-_107218786
|
28.191
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr10_-_64578926
|
25.667
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr10_-_64576123
|
19.593
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64576074
|
18.180
|
|
EGR2
|
early growth response 2
|
|
chrX_-_106960028
|
16.518
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_27860927
|
15.203
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chrX_-_106960268
|
14.868
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_34642651
|
13.862
|
|
EHF
|
ets homologous factor
|
|
chr6_-_11232883
|
13.504
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11232900
|
13.161
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr11_+_34642587
|
13.147
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr1_+_68151092
|
13.011
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr6_-_11232901
|
12.973
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_68150858
|
12.905
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr15_-_41166336
|
11.965
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr2_-_207082626
|
11.802
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr5_+_145317297
|
11.011
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr2_-_239197091
|
10.138
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr7_+_37779995
|
9.534
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr10_+_123922940
|
9.141
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr6_+_26124372
|
8.865
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr6_+_27100784
|
8.742
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr10_+_123923104
|
8.540
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr1_+_61547625
|
7.995
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_37381920
|
7.989
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr17_-_7832752
|
7.876
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr5_-_2751768
|
7.735
|
NM_001134222
NM_033267
|
IRX2
|
iroquois homeobox 2
|
|
chr19_-_18392048
|
7.543
|
|
JUND
|
jun D proto-oncogene
|
|
chr11_+_537521
|
7.497
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr7_+_100183926
|
7.448
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr4_-_47840058
|
7.401
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr6_-_27782517
|
7.399
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr1_-_228645517
|
7.239
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr12_+_53442732
|
7.047
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr20_+_33292117
|
7.032
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr17_-_19651586
|
6.969
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr6_-_26216871
|
6.767
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr19_-_18392436
|
6.737
|
|
JUND
|
jun D proto-oncogene
|
|
chr6_-_35656610
|
6.710
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr3_+_132036210
|
6.689
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr1_+_18807423
|
6.517
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr6_-_35656677
|
6.462
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chrX_-_15402469
|
6.432
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr7_-_100183722
|
6.352
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr19_-_18392413
|
6.303
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr13_+_52027435
|
6.216
|
|
|
|
|
chr3_-_87040268
|
6.075
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_+_26183957
|
6.035
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr2_+_152214105
|
6.023
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr16_-_74808699
|
5.994
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr16_+_85645014
|
5.984
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr16_-_74808474
|
5.937
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr12_-_11091805
|
5.804
|
NM_023922
|
TAS2R14
|
taste receptor, type 2, member 14
|
|
chr9_+_109686436
|
5.800
|
|
ZNF462
|
zinc finger protein 462
|
|
chr17_-_8063946
|
5.774
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr12_+_53442972
|
5.740
|
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr16_-_30107492
|
5.732
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr5_+_2752261
|
5.729
|
NM_178569
|
C5orf38
|
chromosome 5 open reading frame 38
|
|
chr3_-_87040246
|
5.620
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr11_+_112047088
|
5.618
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr9_+_136325086
|
5.521
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr6_+_27114860
|
5.349
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr3_+_132036276
|
5.327
|
|
ACPP
|
acid phosphatase, prostate
|
|
chr18_+_44526786
|
5.325
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr2_+_211458078
|
4.790
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr1_+_228645807
|
4.759
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr7_+_18535351
|
4.677
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr14_-_107199126
|
4.640
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr1_-_149859465
|
4.611
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr17_+_72983723
|
4.588
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr6_+_10585978
|
4.514
|
NM_145655
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr16_+_30107742
|
4.355
|
|
|
|
|
chr19_-_44123732
|
4.286
|
|
ZNF428
|
zinc finger protein 428
|
|
chr13_+_97794050
|
4.285
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr6_-_27100528
|
4.207
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr19_-_50143350
|
4.196
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr6_+_43139201
|
4.125
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr19_+_56111600
|
4.123
|
NM_153219
|
ZNF524
|
zinc finger protein 524
|
|
chr12_+_14518608
|
3.958
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr6_+_27833033
|
3.958
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr10_-_98031154
|
3.837
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_-_224467142
|
3.774
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr11_+_27062508
|
3.702
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr11_+_112046207
|
3.677
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr17_+_43213013
|
3.620
|
NM_001135705
NM_001135706
NM_001135707
NM_024722
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr14_-_106791054
|
3.605
|
|
IGHV4-31
IGHV3-30
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-30
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr7_-_100183753
|
3.552
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr19_-_44124013
|
3.531
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr5_+_60628085
|
3.464
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr17_+_20059192
|
3.444
|
NM_001033554
NM_001033555
NM_001243438
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr1_-_186649421
|
3.436
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr1_+_61547388
|
3.431
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chrX_-_117119282
|
3.380
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr8_-_25282555
|
3.378
|
NM_000825
NM_001083111
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone)
|
|
chr4_-_186733372
|
3.351
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_26033795
|
3.351
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr19_-_47164289
|
3.283
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr10_-_97321094
|
3.232
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr2_+_97426571
|
3.223
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr6_+_26217145
|
3.154
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr9_+_129677046
|
3.128
|
NM_001190729
NM_001190730
NM_014636
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr4_-_40517913
|
3.108
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr6_+_26251826
|
3.063
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr7_+_141490016
|
3.004
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chr12_-_57443858
|
2.934
|
NM_005379
|
MYO1A
|
myosin IA
|
|
chr1_+_6052758
|
2.841
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chrX_-_55020510
|
2.795
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr14_-_24553817
|
2.794
|
NM_006177
|
NRL
|
neural retina leucine zipper
|
|
chr6_+_148663728
|
2.760
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr3_-_48594209
|
2.712
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr7_+_90338711
|
2.695
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_+_148663953
|
2.660
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_206730456
|
2.578
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr4_-_18023358
|
2.568
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr20_+_34556528
|
2.567
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr19_+_17337012
|
2.530
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr12_-_56359794
|
2.508
|
NM_001200053
NM_001200054
|
PMEL
|
premelanosome protein
|
|
chr1_+_6052699
|
2.443
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr12_-_6484557
|
2.439
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr22_-_24096551
|
2.405
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chrX_-_80457440
|
2.399
|
NM_030763
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr6_+_27861202
|
2.392
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr6_+_43139190
|
2.370
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr12_-_6484389
|
2.366
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr22_+_44319685
|
2.359
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr1_-_149858114
|
2.343
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr20_-_40246991
|
2.315
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr1_+_206730497
|
2.271
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr3_-_172241060
|
2.257
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr22_-_36556781
|
2.256
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr19_+_3572958
|
2.223
|
|
HMG20B
|
high mobility group 20B
|
|
chr12_-_122240827
|
2.180
|
|
|
|
|
chrX_-_131547536
|
2.177
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr18_-_52988916
|
2.160
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr22_+_44319674
|
2.126
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr19_-_19144304
|
2.050
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr22_+_44319618
|
2.047
|
NM_025225
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr2_-_113594325
|
2.022
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr10_-_62332666
|
2.011
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr7_-_27153490
|
1.971
|
|
HOXA3
|
homeobox A3
|
|
chr6_+_27782793
|
1.970
|
NM_003521
|
HIST1H2BM
|
histone cluster 1, H2bm
|
|
chr19_-_19144262
|
1.923
|
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr8_+_42396297
|
1.870
|
NM_001135676
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr11_+_120107348
|
1.851
|
NM_001244682
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr6_+_43138919
|
1.850
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr5_-_39219578
|
1.844
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr19_-_893184
|
1.843
|
|
MED16
|
mediator complex subunit 16
|
|
chr5_+_173472692
|
1.827
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr19_+_1269244
|
1.771
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr15_-_89456592
|
1.771
|
NM_001114614
NM_005928
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr3_+_111758464
|
1.766
|
NM_001042575
|
TMPRSS7
|
transmembrane protease, serine 7
|
|
chr17_+_80317122
|
1.761
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr7_+_94539209
|
1.749
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr3_-_121379701
|
1.746
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr15_-_89456549
|
1.739
|
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr19_-_19144326
|
1.725
|
NM_001017392
NM_014884
|
SUGP2
|
SURP and G patch domain containing 2
|
|
chr17_+_9745847
|
1.708
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr18_+_76740274
|
1.693
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr6_+_139094656
|
1.679
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr3_-_123603144
|
1.667
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr5_-_176935877
|
1.641
|
|
DOK3
|
docking protein 3
|
|
chr19_+_58907456
|
1.636
|
NM_001195135
|
LOC646862
|
uncharacterized LOC646862
|
|
chrX_-_65859107
|
1.618
|
NM_001199687
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr2_+_202047620
|
1.612
|
NM_001206524
NM_001206542
NM_001230
NM_032974
NM_032976
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr14_-_106994001
|
1.598
|
|
IGHV3-48
IGH@
IGHA1
IGHM
|
immunoglobulin heavy variable 3-48
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr19_+_3572903
|
1.594
|
NM_006339
|
HMG20B
|
high mobility group 20B
|
|
chr17_-_56494930
|
1.587
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr5_-_175964824
|
1.582
|
|
RNF44
|
ring finger protein 44
|
|
chr19_+_3572950
|
1.570
|
|
HMG20B
|
high mobility group 20B
|
|
chr1_-_149814264
|
1.542
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr2_-_172750724
|
1.534
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr19_-_40931880
|
1.525
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr15_-_89089792
|
1.500
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr16_-_88752788
|
1.497
|
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr19_-_893215
|
1.496
|
NM_005481
|
MED16
|
mediator complex subunit 16
|
|
chr19_-_19281062
|
1.488
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr19_-_19281090
|
1.478
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chrX_-_102347950
|
1.449
|
NM_022052
|
NXF3
|
nuclear RNA export factor 3
|
|
chr6_-_26124130
|
1.445
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr12_-_57328044
|
1.436
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr6_+_26158332
|
1.423
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr17_-_67138013
|
1.421
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr6_+_27215479
|
1.419
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr1_+_33722158
|
1.395
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr3_-_149086884
|
1.390
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chrM_+_5769
|
1.374
|
|
|
|
|
chr1_+_149822627
|
1.367
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr10_-_123356158
|
1.362
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr16_-_90096308
|
1.360
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr7_+_112120907
|
1.355
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr2_+_207804277
|
1.351
|
NM_173077
|
CPO
|
carboxypeptidase O
|
|
chr20_-_49547451
|
1.347
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr22_+_24033047
|
1.344
|
NM_153615
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr16_+_53164784
|
1.320
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr4_-_152682092
|
1.311
|
NM_004564
|
PET112
|
PET112 homolog (yeast)
|
|
chr2_+_170683900
|
1.304
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr10_+_32856650
|
1.294
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|