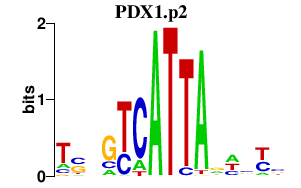
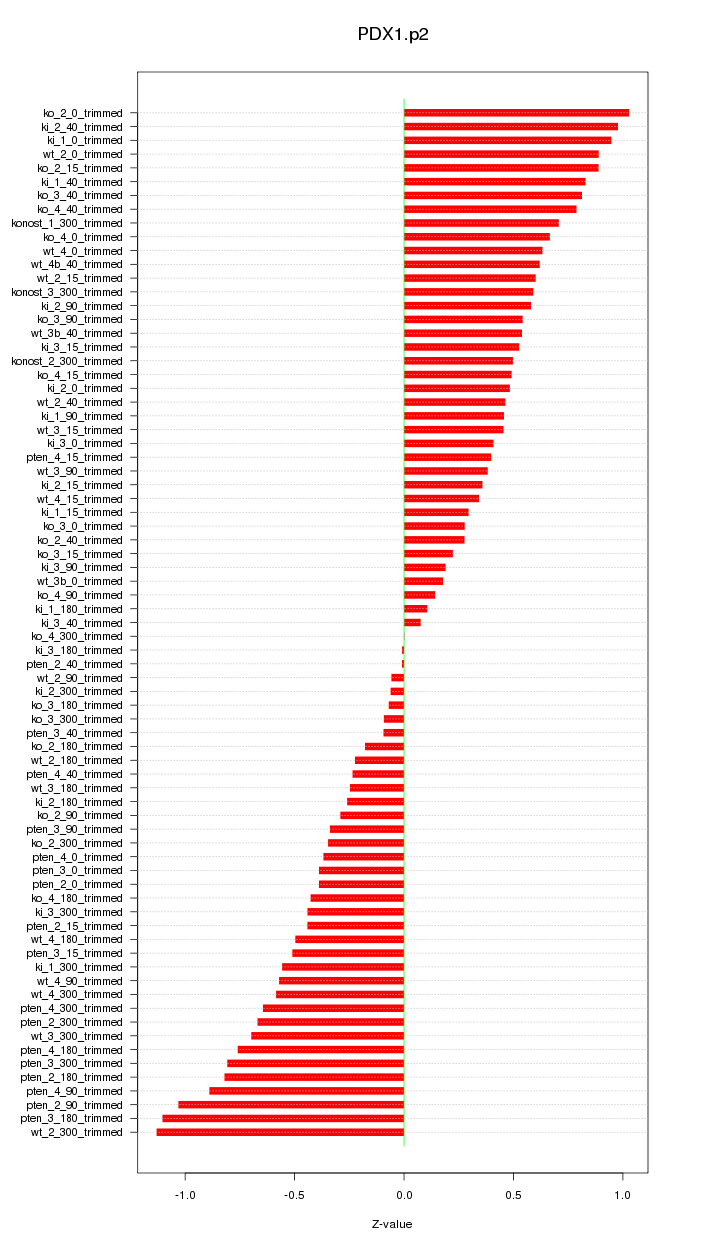
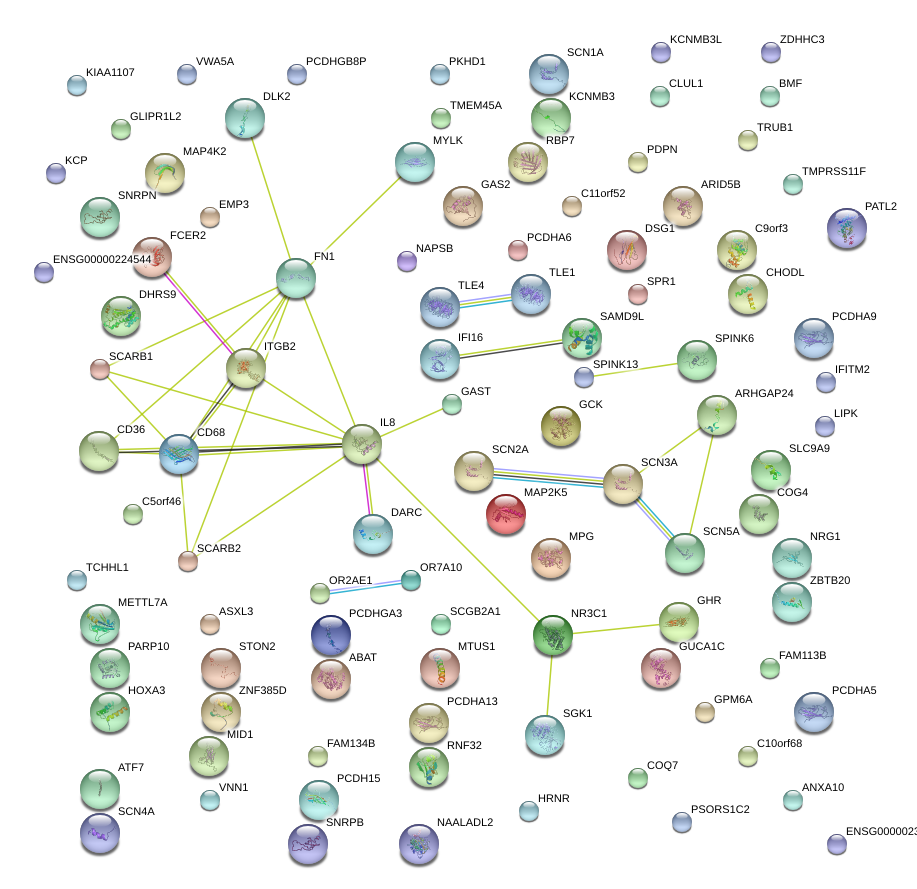
|
chr15_-_40398286
|
5.087
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr2_+_113735595
|
3.871
|
NM_019618
|
IL36G
|
interleukin 36, gamma
|
|
chr4_+_86396251
|
3.335
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_+_100238063
|
3.242
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr6_-_133035180
|
3.011
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr7_-_92777657
|
2.841
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr6_-_31107110
|
2.727
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr15_+_67841341
|
2.137
|
NM_001206804
|
MAP2K5
|
mitogen-activated protein kinase kinase 5
|
|
chr10_+_63808968
|
2.080
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr15_-_40398573
|
1.904
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr15_-_44969085
|
1.778
|
NM_001145112
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast)
|
|
chr6_-_134498973
|
1.694
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_158979681
|
1.505
|
NM_001206567
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr6_-_43423785
|
1.492
|
NM_206539
|
DLK2
|
delta-like 2 homolog (Drosophila)
|
|
chr3_-_108672547
|
1.488
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr15_+_25281105
|
1.485
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr7_+_80267972
|
1.402
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr2_-_201967702
|
1.384
|
|
|
|
|
chr3_+_174577069
|
1.267
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr11_+_22696381
|
1.093
|
|
GAS2
|
growth arrest-specific 2
|
|
chr4_-_68995586
|
1.077
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr3_-_143567163
|
1.076
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr19_+_48828627
|
1.027
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr16_-_70557430
|
1.009
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr5_+_140201221
|
0.995
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr12_+_47610051
|
0.988
|
NM_138371
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr4_+_74606222
|
0.983
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr4_-_176733413
|
0.941
|
|
GPM6A
|
glycoprotein M6A
|
|
chr11_+_111789600
|
0.821
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr1_+_10057254
|
0.785
|
NM_052960
|
RBP7
|
retinol binding protein 7, cellular
|
|
chr9_+_82187469
|
0.775
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr5_+_42565562
|
0.725
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr14_-_81864726
|
0.710
|
NM_033104
|
STON2
|
stonin 2
|
|
chr21_-_46348633
|
0.691
|
NM_001127491
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr14_-_77889231
|
0.686
|
NM_001113475
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1
|
|
chr19_-_36019194
|
0.686
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr5_+_147582338
|
0.668
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr11_+_123986103
|
0.661
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr1_+_92632608
|
0.574
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr5_+_147648374
|
0.572
|
NM_001040129
|
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative)
|
|
chr16_+_28505920
|
0.558
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr8_-_17580024
|
0.548
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr4_-_176733378
|
0.543
|
|
GPM6A
|
glycoprotein M6A
|
|
chr12_+_51318515
|
0.536
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr16_+_19078916
|
0.535
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr16_+_19078958
|
0.531
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr10_+_90484300
|
0.523
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr2_-_216259197
|
0.514
|
|
FN1
|
fibronectin 1
|
|
chr8_-_17579729
|
0.505
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr1_-_29309092
|
0.488
|
|
|
|
|
chr7_+_80275967
|
0.486
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr8_-_17555158
|
0.480
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_8814567
|
0.477
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr9_+_82187599
|
0.476
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr18_+_28898051
|
0.475
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr11_+_22696318
|
0.462
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr2_+_169921298
|
0.457
|
NM_005771
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr12_+_75784878
|
0.443
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chrX_-_10645726
|
0.424
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_+_47610244
|
0.401
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr12_+_31800093
|
0.399
|
NM_001135864
|
METTL20
|
methyltransferase like 20
|
|
chr16_+_19079220
|
0.397
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr5_+_140718353
|
0.396
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr17_+_7482938
|
0.391
|
|
CD68
|
CD68 molecule
|
|
chr10_+_32856650
|
0.389
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr3_-_123339423
|
0.373
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr17_+_7482980
|
0.369
|
|
CD68
|
CD68 molecule
|
|
chr7_-_44228880
|
0.348
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr3_-_21792815
|
0.347
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr17_+_7482961
|
0.340
|
|
CD68
|
CD68 molecule
|
|
chr2_+_169926084
|
0.333
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_+_159175048
|
0.332
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr8_-_145060594
|
0.326
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr1_-_152061529
|
0.323
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr4_+_169013687
|
0.317
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr7_-_128543379
|
0.310
|
|
KCP
|
kielin/chordin-like protein
|
|
chr1_-_152196668
|
0.302
|
NM_001009931
|
HRNR
|
hornerin
|
|
chr5_+_140235594
|
0.302
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr9_+_97766414
|
0.302
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr3_-_45017600
|
0.301
|
NM_001135179
NM_016598
|
ZDHHC3
|
zinc finger, DHHC-type containing 3
|
|
chr18_+_616671
|
0.288
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr10_-_56560947
|
0.266
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr5_-_147286064
|
0.261
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr4_-_176733901
|
0.251
|
|
GPM6A
|
glycoprotein M6A
|
|
chr21_+_19617145
|
0.241
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr5_-_16509106
|
0.240
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr12_-_54020081
|
0.226
|
NM_001130060
NM_001206682
NM_006856
|
ATF7
|
activating transcription factor 7
|
|
chr12_-_54019633
|
0.222
|
NM_001206683
|
ATF7
|
activating transcription factor 7
|
|
chr5_+_140227356
|
0.208
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr10_+_116697914
|
0.203
|
NM_139169
|
TRUB1
|
TruB pseudouridine (psi) synthase homolog 1 (E. coli)
|
|
chr19_-_7766978
|
0.196
|
NM_001220500
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr17_+_7482804
|
0.188
|
NM_001251
NM_001040059
|
CD68
|
CD68 molecule
|
|
chr3_-_178969644
|
0.184
|
NM_171830
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr12_+_75784849
|
0.178
|
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr19_-_50847957
|
0.163
|
|
NAPSB
|
napsin B aspartic peptidase pseudogene
|
|
chr18_+_31319154
|
0.162
|
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr8_+_32579350
|
0.155
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr7_-_99474655
|
0.152
|
NM_001005276
|
OR2AE1
|
olfactory receptor, family 2, subfamily AE, member 1
|
|
chr2_-_166060552
|
0.145
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr17_+_39555993
|
0.145
|
|
GAST
|
gastrin
|
|
chr3_-_114343791
|
0.140
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_47923271
|
0.140
|
NM_001242791
|
FLJ45513
|
uncharacterized LOC729220
|
|
chr6_-_51952368
|
0.138
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr9_+_97521930
|
0.137
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr7_+_156435670
|
0.137
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chr1_+_13911966
|
0.135
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr7_-_27166638
|
0.134
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr19_-_14952688
|
0.129
|
NM_001005190
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10
|
|
chr5_-_142815076
|
0.124
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr11_+_61976139
|
0.114
|
NM_002407
|
SCGB2A1
|
secretoglobin, family 2A, member 1
|
|
chr17_+_7960201
|
0.108
|
|
|
|
|
chr6_+_46761093
|
0.106
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr16_-_48269087
|
0.102
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr17_-_7082654
|
0.098
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr10_+_88730497
|
0.098
|
NM_133447
|
AGAP11
|
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
|
|
chr3_-_151034514
|
0.097
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr3_-_137832321
|
0.094
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr6_+_131571298
|
0.091
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr1_+_43735664
|
0.089
|
NM_144626
|
TMEM125
|
transmembrane protein 125
|
|
chr17_+_38119225
|
0.089
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr3_+_197879236
|
0.088
|
NM_001145248
|
FAM157A
|
family with sequence similarity 157, member A
|
|
chr7_+_129984629
|
0.087
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr3_-_114478117
|
0.087
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_76382458
|
0.085
|
|
LOC728723
|
uncharacterized LOC728723
|
|
chr16_+_57728711
|
0.081
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr1_+_84768333
|
0.080
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr12_-_14849427
|
0.079
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr12_+_15475190
|
0.073
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr6_+_72922513
|
0.071
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_+_68667280
|
0.071
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr13_-_95131718
|
0.065
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr12_-_9885859
|
0.064
|
NM_001253748
NM_001253750
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr1_-_186417855
|
0.060
|
NM_022576
|
PDC
|
phosducin
|
|
chr13_-_41593491
|
0.055
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr4_+_88896801
|
0.054
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_+_108541544
|
0.050
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr1_+_28764660
|
0.050
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr1_+_36748164
|
0.050
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr6_-_131321868
|
0.048
|
NM_001252660
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr6_+_167704802
|
0.047
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr12_-_103311380
|
0.047
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr6_+_72926144
|
0.046
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_-_57873364
|
0.042
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr16_+_29674282
|
0.042
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr2_+_137748461
|
0.040
|
NM_001080427
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr7_-_44580857
|
0.039
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr3_+_46619075
|
0.038
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr3_+_35683848
|
0.038
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr10_-_62332666
|
0.037
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_163038934
|
0.037
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_+_80838125
|
0.035
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr8_-_27115935
|
0.034
|
|
STMN4
|
stathmin-like 4
|
|
chr10_+_118187381
|
0.034
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr12_+_21525801
|
0.028
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr6_+_42531736
|
0.027
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr12_-_4488707
|
0.026
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr2_+_38893051
|
0.025
|
NM_138801
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr14_+_51955854
|
0.023
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr3_+_45017749
|
0.018
|
|
EXOSC7
|
exosome component 7
|
|
chr7_-_87342569
|
0.018
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr4_-_24981769
|
0.017
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr6_+_149887542
|
0.017
|
|
C6orf72
|
chromosome 6 open reading frame 72
|
|
chr3_+_45017729
|
0.016
|
NM_015004
|
EXOSC7
|
exosome component 7
|
|
chr6_+_144612872
|
0.016
|
NM_007124
|
UTRN
|
utrophin
|
|
chr6_-_62995852
|
0.014
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr21_+_31768294
|
0.014
|
NM_181599
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chr8_+_92261515
|
0.014
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr12_-_11214892
|
0.013
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr9_+_141106636
|
0.008
|
NM_001145249
|
FAM157B
|
family with sequence similarity 157, member B
|
|
chr6_-_131291480
|
0.008
|
NM_001199389
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chrX_-_76814217
|
0.005
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr3_-_167191808
|
0.005
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chrX_+_49091931
|
0.004
|
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr2_+_38893238
|
0.003
|
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr21_-_31538970
|
0.002
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr1_+_84874033
|
0.001
|
NM_058248
|
DNASE2B
|
deoxyribonuclease II beta
|