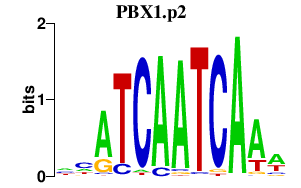
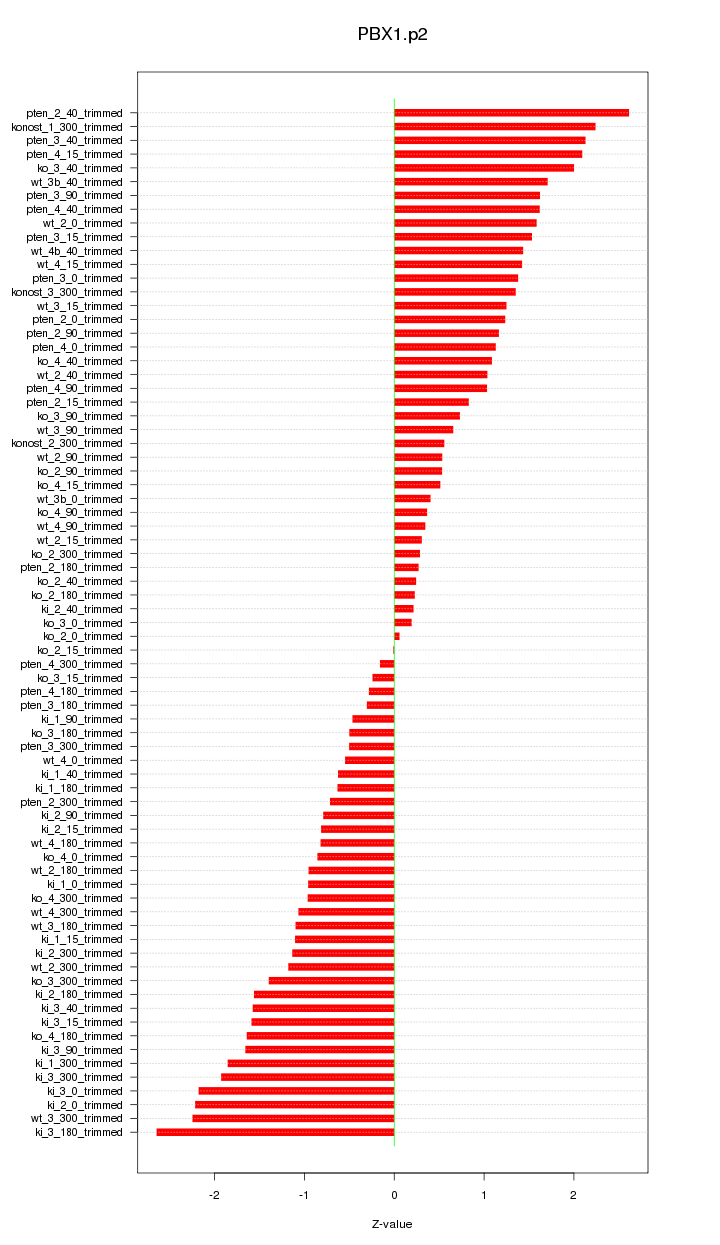
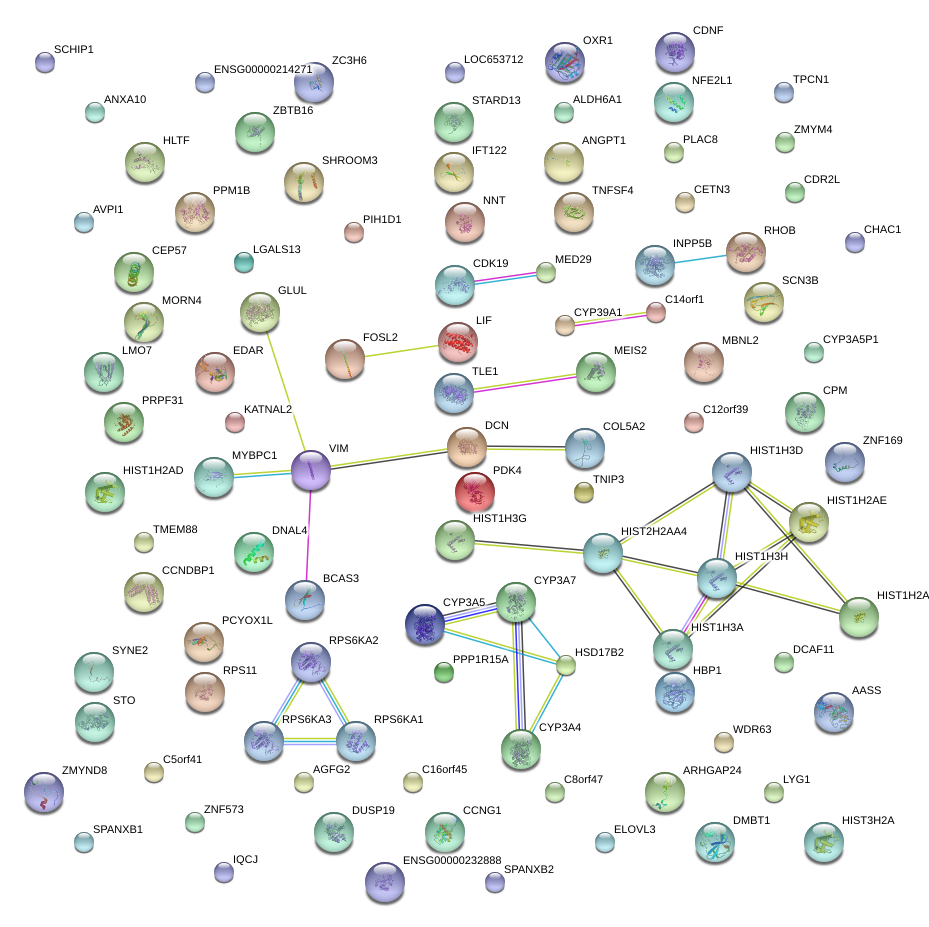
|
chr16_+_15596122
|
13.538
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr12_+_101988746
|
12.861
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr1_+_85527992
|
11.887
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr22_-_30642683
|
10.007
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_-_29309092
|
8.380
|
|
|
|
|
chr22_-_39190103
|
6.950
|
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr19_+_49375677
|
6.869
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr19_+_49375679
|
6.838
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr6_-_167040700
|
6.808
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr7_-_95225802
|
6.699
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr19_+_49375638
|
6.304
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr2_-_99917638
|
6.148
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr6_-_167040506
|
6.032
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_+_124320180
|
6.010
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr1_-_182360889
|
5.530
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr14_-_74551073
|
5.504
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr12_-_91573264
|
5.493
|
NM_133503
|
DCN
|
decorin
|
|
chr6_-_167040659
|
5.231
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr8_-_108510094
|
5.224
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr13_+_97928414
|
5.218
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_+_106809405
|
5.165
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr22_-_39190146
|
5.110
|
NM_005740
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr17_+_46125675
|
4.703
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr6_-_26199439
|
4.662
|
NM_021065
NM_003530
|
HIST1H2AD
HIST1H3D
|
histone cluster 1, H2ad
histone cluster 1, H3d
|
|
chr17_+_46125724
|
4.597
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr17_+_46125725
|
4.455
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr5_+_148737569
|
4.436
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chrX_+_140084755
|
4.383
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chr14_-_74551159
|
4.357
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr13_-_33859835
|
4.294
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_+_82068830
|
4.140
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr6_-_111136616
|
4.051
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr2_+_28616346
|
4.023
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_-_109605668
|
4.018
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr13_+_97928196
|
3.983
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr19_-_46220589
|
3.899
|
|
|
|
|
chr18_+_44526786
|
3.831
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr12_+_113659216
|
3.659
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr11_+_95523608
|
3.651
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr4_+_86749045
|
3.583
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_+_95523809
|
3.566
|
|
CEP57
|
centrosomal protein 57kDa
|
|
chr20_-_45976626
|
3.565
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr11_-_123525300
|
3.518
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr12_-_69357019
|
3.508
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr9_-_84304378
|
3.489
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr5_+_162864571
|
3.477
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr15_-_37391893
|
3.446
|
|
MEIS2
|
Meis homeobox 2
|
|
chr14_+_64680858
|
3.428
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr2_-_190044330
|
3.407
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_+_159557649
|
3.353
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_+_82068910
|
3.335
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr3_+_159557745
|
3.208
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_113659287
|
3.196
|
|
TPCN1
|
two pore segment channel 1
|
|
chr1_-_38397417
|
3.182
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr10_+_103986142
|
3.171
|
NM_152310
|
ELOVL3
|
ELOVL fatty acid elongase 3
|
|
chr3_+_129158949
|
3.028
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr14_+_24584321
|
3.025
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr12_+_113659275
|
2.984
|
|
TPCN1
|
two pore segment channel 1
|
|
chr2_+_44395983
|
2.943
|
NM_001033556
NM_001033557
NM_002706
NM_177968
NM_177969
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr14_-_76127158
|
2.928
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr12_+_21679190
|
2.910
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr8_+_99076749
|
2.908
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr1_-_182360400
|
2.845
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_-_89705477
|
2.811
|
NM_004365
|
CETN3
|
centrin, EF-hand protein, 3
|
|
chr5_+_43602738
|
2.782
|
NM_012343
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr17_+_72983723
|
2.745
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr4_-_84035888
|
2.702
|
|
PLAC8
|
placenta-specific 8
|
|
chr3_-_148804122
|
2.658
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr10_-_14879967
|
2.656
|
NM_001029954
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr2_+_200323088
|
2.589
|
|
|
|
|
chr11_+_113931228
|
2.584
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr19_+_54619149
|
2.577
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr3_+_129159150
|
2.555
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr7_+_100136784
|
2.550
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr3_+_129159075
|
2.548
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr5_+_172483285
|
2.534
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr15_-_37393364
|
2.444
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr13_+_76194569
|
2.429
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr19_+_49999633
|
2.401
|
NM_001015
|
RPS11
|
ribosomal protein S11
|
|
chr19_+_49999682
|
2.393
|
|
RPS11
|
ribosomal protein S11
|
|
chr17_+_7758380
|
2.380
|
NM_203411
|
TMEM88
|
transmembrane protein 88
|
|
chr10_+_17270257
|
2.363
|
NM_003380
|
VIM
|
vimentin
|
|
chr2_+_28616360
|
2.338
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr15_+_43477315
|
2.332
|
NM_012142
|
CCNDBP1
|
cyclin D-type binding-protein 1
|
|
chr19_+_39882018
|
2.316
|
|
MED29
|
mediator complex subunit 29
|
|
chr2_+_113033146
|
2.315
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr1_-_173176314
|
2.300
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr19_+_39881950
|
2.281
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr17_+_58755158
|
2.254
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr19_+_49999687
|
2.254
|
|
RPS11
|
ribosomal protein S11
|
|
chr2_+_44396061
|
2.249
|
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr5_+_43603287
|
2.239
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr7_-_121784236
|
2.219
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr10_-_99446899
|
2.214
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr8_+_107738239
|
2.211
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr15_+_41245635
|
2.206
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr6_-_46620229
|
2.201
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr5_-_151150939
|
2.193
|
|
LOC100652758
|
uncharacterized LOC100652758
|
|
chr2_+_113033184
|
2.187
|
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr4_+_77356252
|
2.156
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr14_+_24583979
|
2.152
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr4_+_169013687
|
2.150
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr19_-_49955025
|
2.147
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr4_-_122137781
|
2.140
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr3_-_12586962
|
2.132
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr5_+_176562084
|
2.130
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr7_-_99332715
|
2.115
|
NM_000765
|
CYP3A7
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 7
cytochrome P450, family 3, subfamily A, polypeptide 5
|
|
chr5_+_43603312
|
2.084
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr1_+_35851101
|
2.061
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr6_-_26271588
|
2.060
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr6_+_26124372
|
2.044
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr2_+_20646831
|
2.040
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr10_-_99393851
|
2.008
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr2_+_183943511
|
2.005
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr12_+_51442081
|
2.003
|
NM_001243689
NM_015416
|
LETMD1
|
LETM1 domain containing 1
|
|
chr3_+_193853937
|
1.957
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_+_193853930
|
1.943
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr4_-_84035908
|
1.941
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr14_+_24584009
|
1.936
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr5_+_43603216
|
1.920
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr3_+_129159089
|
1.913
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr16_+_67311412
|
1.901
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr2_-_96482382
|
1.895
|
|
LINC00342
|
long intergenic non-protein coding RNA 342
|
|
chr1_-_19229016
|
1.891
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr3_+_52570620
|
1.874
|
NM_001124767
|
C3orf78
|
chromosome 3 open reading frame 78
|
|
chr10_+_63661395
|
1.853
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr14_+_74551655
|
1.851
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr1_+_145663156
|
1.850
|
|
RNF115
|
ring finger protein 115
|
|
chr6_+_31707724
|
1.824
|
NM_002441
NM_025259
NM_172165
NM_172166
|
MSH5-C6orf26
MSH5
|
MSH5-C6orf26 readthrough (non-protein coding)
mutS homolog 5 (E. coli)
|
|
chr3_+_111451326
|
1.819
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr14_+_24583905
|
1.817
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_-_200379165
|
1.814
|
NM_012482
|
ZNF281
|
zinc finger protein 281
|
|
chr1_-_147610080
|
1.796
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr3_+_129159138
|
1.791
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr12_-_120315074
|
1.785
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr17_-_45973142
|
1.755
|
|
|
|
|
chr22_+_35695872
|
1.744
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr19_-_11266451
|
1.735
|
NM_182513
|
SPC24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr16_+_67927170
|
1.729
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr17_+_57408907
|
1.704
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr19_+_54619114
|
1.685
|
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr1_-_146068244
|
1.683
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr1_-_228645517
|
1.676
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr10_+_5005685
|
1.651
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr4_+_71384289
|
1.605
|
NM_212557
|
AMTN
|
amelotin
|
|
chr2_-_145277879
|
1.579
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr9_-_95298251
|
1.565
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr11_+_19372270
|
1.559
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr3_+_136581074
|
1.533
|
|
NCK1
|
NCK adaptor protein 1
|
|
chr6_+_26225382
|
1.519
|
NM_003532
|
HIST1H3E
|
histone cluster 1, H3e
|
|
chr4_-_1722978
|
1.514
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr11_+_122753390
|
1.510
|
NM_024806
NM_199124
|
C11orf63
|
chromosome 11 open reading frame 63
|
|
chr19_+_54704659
|
1.499
|
NM_001013
|
RPS9
|
ribosomal protein S9
|
|
chr6_+_46620631
|
1.496
|
NM_001204051
NM_001204052
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr10_+_63661012
|
1.496
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr17_-_73663221
|
1.486
|
NM_001003715
NM_001003716
NM_004259
|
RECQL5
|
RecQ protein-like 5
|
|
chr19_+_13906207
|
1.478
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr3_-_129158675
|
1.477
|
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chr10_+_5238756
|
1.471
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr21_-_47648715
|
1.470
|
NM_001001438
NM_001145436
NM_001145437
NM_002340
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr1_-_221915426
|
1.453
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr2_+_183943286
|
1.439
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr12_-_120638894
|
1.425
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr16_+_11439279
|
1.425
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr4_-_141677470
|
1.421
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr17_-_41977966
|
1.420
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr12_-_120638909
|
1.406
|
|
RPLP0
|
ribosomal protein, large, P0
|
|
chr1_+_151763174
|
1.402
|
|
|
|
|
chr7_-_7680048
|
1.384
|
|
RPA3
|
replication protein A3, 14kDa
|
|
chr7_-_27142301
|
1.378
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr10_+_76586299
|
1.367
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr3_-_129158779
|
1.361
|
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chr22_-_30722863
|
1.359
|
NM_001204240
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr14_+_76127550
|
1.358
|
NM_015072
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr16_+_11439311
|
1.356
|
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr8_+_18079176
|
1.354
|
NM_001160174
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr14_+_76127592
|
1.350
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr10_+_13142055
|
1.346
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr16_+_838645
|
1.339
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr22_+_35695792
|
1.336
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr19_-_39881503
|
1.316
|
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr17_-_44439083
|
1.313
|
NM_001039083
NM_001103154
NM_001113738
NM_016632
|
ARL17B
ARL17A
|
ADP-ribosylation factor-like 17B
ADP-ribosylation factor-like 17A
|
|
chr17_-_38821286
|
1.307
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr2_-_200322699
|
1.300
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr1_-_182354649
|
1.294
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr15_+_59908724
|
1.290
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr22_+_29138018
|
1.287
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr15_+_93443559
|
1.287
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr14_+_64971504
|
1.285
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr6_-_31830593
|
1.281
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr15_+_93443418
|
1.280
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_-_174255454
|
1.269
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr19_+_1270926
|
1.267
|
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr9_-_14313509
|
1.258
|
|
NFIB
|
nuclear factor I/B
|
|
chr20_-_44563573
|
1.232
|
|
FLJ40606
|
uncharacterized LOC643549
|
|
chr10_+_111765725
|
1.228
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr15_-_72612247
|
1.222
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr14_+_76127656
|
1.215
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chrM_+_9725
|
1.209
|
|
|
|
|
chr1_+_151227209
|
1.209
|
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|