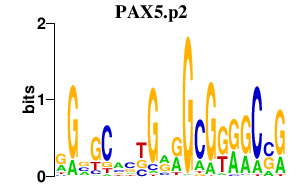
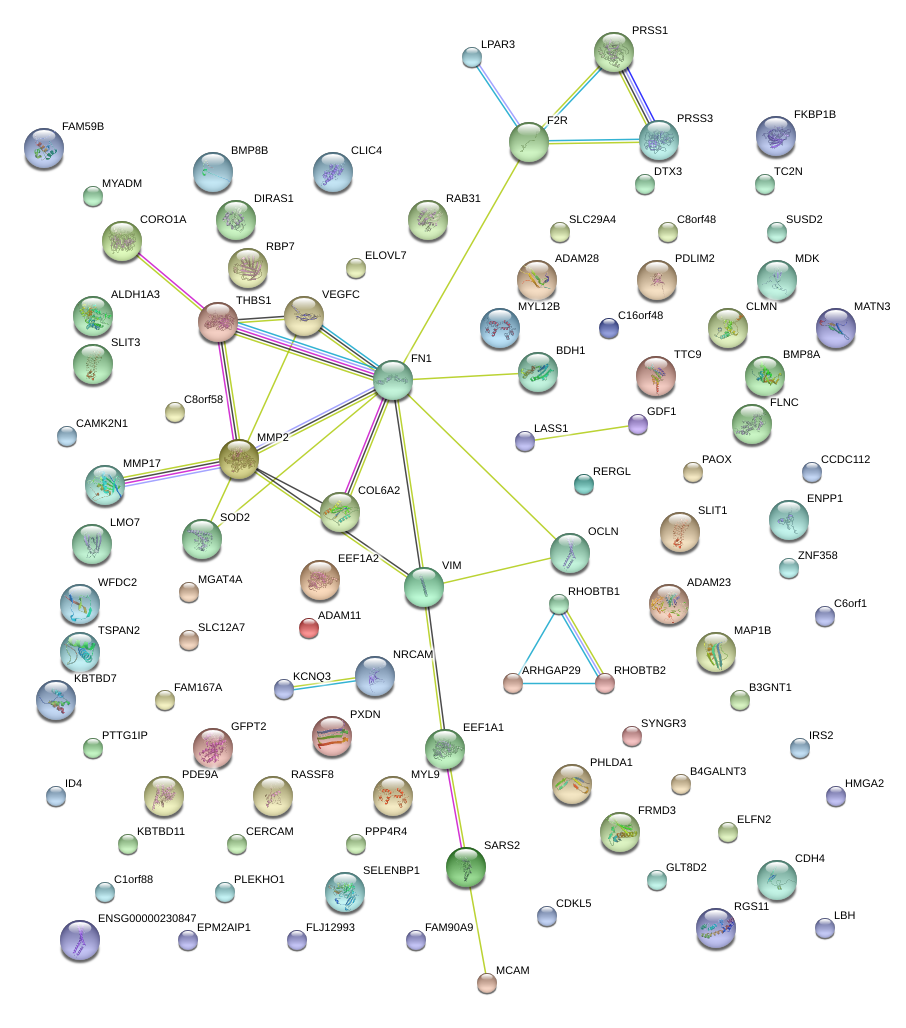
|
chr16_-_325867
|
21.984
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr1_-_85358851
|
15.744
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr15_+_101420004
|
11.006
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr7_+_128470462
|
10.789
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr20_-_62130390
|
10.527
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr9_-_86153048
|
10.494
|
NM_001244959
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr9_+_33750463
|
10.340
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr5_-_168727715
|
9.679
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr15_+_101420032
|
8.564
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr2_-_216300770
|
8.402
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr7_-_108096693
|
7.923
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr10_+_135192694
|
7.830
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr8_+_1922030
|
7.553
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr5_-_1112106
|
7.408
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr1_+_10057254
|
7.148
|
NM_052960
|
RBP7
|
retinol binding protein 7, cellular
|
|
chr14_-_95786095
|
7.114
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr11_+_46403260
|
6.470
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr16_-_67700571
|
6.300
|
NM_032140
|
C16orf48
|
chromosome 16 open reading frame 48
|
|
chr5_-_60140041
|
6.091
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL fatty acid elongase 7
|
|
chr2_-_216300444
|
5.900
|
|
FN1
|
fibronectin 1
|
|
chr20_+_44098392
|
5.832
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr2_-_99347430
|
5.719
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr4_-_177713664
|
5.522
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr11_+_46403205
|
5.470
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr8_-_91997437
|
5.460
|
NM_001190972
|
LOC100127983
|
uncharacterized LOC100127983
|
|
chr1_-_115632047
|
5.373
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr21_-_46293598
|
5.170
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr13_-_110438896
|
5.110
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr2_+_24272627
|
5.092
|
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr3_-_197282805
|
5.076
|
NM_004051
NM_203314
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr16_+_2039945
|
5.069
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr21_-_46293532
|
5.065
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr19_-_2721337
|
5.021
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr21_-_46293561
|
5.008
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr18_+_9708187
|
4.973
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_+_68788387
|
4.972
|
NM_001205254
|
OCLN
|
occludin
|
|
chr8_-_133493003
|
4.940
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr19_-_19006868
|
4.914
|
NM_021267
NM_198207
NM_001492
|
CERS1
GDF1
|
ceramide synthase 1
growth differentiation factor 1
|
|
chr20_+_35169897
|
4.866
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr21_-_46293609
|
4.857
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr2_+_30454396
|
4.776
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr20_+_35169876
|
4.638
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr2_-_1748240
|
4.603
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr8_+_13424351
|
4.543
|
NM_001007090
|
C8orf48
|
chromosome 8 open reading frame 48
|
|
chr22_+_24577443
|
4.515
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr19_+_7580987
|
4.513
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr14_+_94640648
|
4.465
|
NM_020958
NM_058237
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4
|
|
chr12_+_66218879
|
4.451
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr18_+_9708322
|
4.450
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_+_76011780
|
4.413
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr12_+_57998600
|
4.405
|
NM_178502
|
DTX3
|
deltex homolog 3 (Drosophila)
|
|
chr8_+_22457098
|
4.392
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr13_-_41768533
|
4.372
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr20_+_59827452
|
4.353
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr2_-_1748285
|
4.350
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr4_-_83483099
|
4.334
|
NM_001080506
|
TMEM150C
|
transmembrane protein 150C
|
|
chr6_+_19837599
|
4.293
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr6_+_132129154
|
4.289
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr2_-_99347588
|
4.270
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr2_-_1748317
|
4.265
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_+_26395959
|
4.254
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr11_-_119187811
|
4.238
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr5_+_71403040
|
4.237
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr16_+_55512801
|
4.216
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_-_1748213
|
4.209
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_111889194
|
4.205
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr15_-_101459410
|
4.200
|
|
|
|
|
chr9_+_131183122
|
4.196
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr1_-_40254521
|
4.194
|
NM_001720
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr10_-_62703822
|
4.173
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr12_-_76425207
|
4.142
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_+_17270469
|
4.072
|
|
VIM
|
vimentin
|
|
chr6_-_160114186
|
4.068
|
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr2_+_24272583
|
3.998
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr21_+_47518012
|
3.976
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_+_132312917
|
3.913
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr14_+_71108373
|
3.861
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr2_-_20212408
|
3.827
|
|
MATN3
|
matrilin 3
|
|
chr2_+_207308280
|
3.827
|
NM_003812
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr22_-_37823503
|
3.783
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr8_-_11324184
|
3.771
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr1_+_150122415
|
3.710
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr15_+_39873276
|
3.693
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chrX_+_18443702
|
3.677
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr17_+_42836546
|
3.666
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr2_+_62423260
|
3.657
|
NM_006577
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2
|
|
chr13_-_41768392
|
3.640
|
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr12_-_76425375
|
3.638
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr14_-_92302612
|
3.636
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr12_+_26111951
|
3.623
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr16_+_30194925
|
3.614
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr22_+_24577417
|
3.609
|
|
SUSD2
|
sushi domain containing 2
|
|
chr16_+_30194730
|
3.597
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr5_-_114632240
|
3.565
|
NM_001040440
|
CCDC112
|
coiled-coil domain containing 112
|
|
chr2_-_20212448
|
3.557
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr11_+_46403279
|
3.551
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr5_-_179780314
|
3.545
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr13_+_76210449
|
3.543
|
|
LMO7
|
LIM domain 7
|
|
chr11_+_46403302
|
3.538
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr1_-_20812271
|
3.532
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr21_+_44073650
|
3.530
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr1_-_40254131
|
3.526
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr1_-_94702916
|
3.442
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_25071967
|
3.440
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr19_+_54371170
|
3.419
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr7_+_5322553
|
3.404
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr12_+_569542
|
3.379
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr18_+_9708269
|
3.370
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr3_+_10206562
|
3.349
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|
|
chr22_+_26825220
|
3.340
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr20_-_36793646
|
3.339
|
NM_004613
NM_198951
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr2_+_97202463
|
3.339
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr8_+_17013459
|
3.334
|
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr1_-_94147201
|
3.320
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr1_-_94703252
|
3.311
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr15_-_52587851
|
3.306
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr7_-_105925329
|
3.297
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr1_-_1535446
|
3.269
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr20_+_62328068
|
3.259
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chrX_-_152864621
|
3.254
|
NM_001130997
NM_152274
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr17_-_15466772
|
3.250
|
NM_001204478
NM_001135036
NM_145301
|
FAM18B2-CDRT4
FAM18B2
|
FAM18B2-CDRT4 readthrough
family with sequence similarity 18, member B2
|
|
chr11_+_46402333
|
3.245
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr1_+_162531229
|
3.225
|
NM_003115
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr17_-_62658311
|
3.217
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr2_+_170590241
|
3.217
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr19_+_54371150
|
3.213
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr15_+_63335086
|
3.208
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr19_+_16187315
|
3.193
|
|
TPM4
|
tropomyosin 4
|
|
chr15_+_76135992
|
3.191
|
|
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2
|
|
chr19_-_46000159
|
3.184
|
NM_005619
NM_206900
|
RTN2
|
reticulon 2
|
|
chr3_+_32280154
|
3.183
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr17_-_29151638
|
3.178
|
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr10_-_62761197
|
3.175
|
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chrX_-_101771641
|
3.147
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chrX_-_153151537
|
3.144
|
|
|
|
|
chr8_+_17013799
|
3.136
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr1_+_16085245
|
3.127
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr19_+_54371125
|
3.107
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr6_+_132129195
|
3.097
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_+_37137882
|
3.071
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chrX_-_152864435
|
3.068
|
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr11_-_125366048
|
3.065
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr3_-_158450205
|
3.053
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr2_+_37571752
|
3.030
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr22_-_38966066
|
3.005
|
NM_007068
|
DMC1
|
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast)
|
|
chr11_-_27494232
|
2.997
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr8_+_17014006
|
2.962
|
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr17_-_62658175
|
2.957
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr20_+_8112762
|
2.953
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr9_+_91606349
|
2.952
|
NM_005226
|
S1PR3
|
sphingosine-1-phosphate receptor 3
|
|
chr18_+_18822168
|
2.947
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chrX_-_38186782
|
2.939
|
NM_000328
NM_001034853
|
RPGR
|
retinitis pigmentosa GTPase regulator
|
|
chr19_-_291168
|
2.934
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr4_+_77227882
|
2.931
|
|
STBD1
|
starch binding domain 1
|
|
chr2_+_210288668
|
2.921
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr9_+_131183225
|
2.921
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr1_-_20812727
|
2.917
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr17_-_41623246
|
2.917
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr10_+_17270257
|
2.899
|
NM_003380
|
VIM
|
vimentin
|
|
chr13_+_98795468
|
2.895
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr6_-_170600165
|
2.894
|
|
|
|
|
chr22_-_20255614
|
2.891
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr6_-_56708444
|
2.890
|
|
DST
|
dystonin
|
|
chr7_+_102074002
|
2.890
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_-_11529948
|
2.886
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr8_+_25042066
|
2.869
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr12_+_93964801
|
2.866
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr11_-_12030622
|
2.856
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr9_+_4490193
|
2.841
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_-_96657538
|
2.835
|
|
ANKRD36C
|
ankyrin repeat domain 36C
|
|
chr17_+_16945766
|
2.831
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr12_+_53491448
|
2.799
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr7_+_89840999
|
2.776
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr12_-_96184346
|
2.766
|
|
NTN4
|
netrin 4
|
|
chr14_+_105992930
|
2.759
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr5_+_52284903
|
2.756
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr9_+_130374466
|
2.756
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr9_-_130331350
|
2.753
|
NM_022833
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr3_+_32280211
|
2.745
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr12_+_27933162
|
2.728
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr1_-_95392501
|
2.728
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr1_+_25071906
|
2.725
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr16_-_90085786
|
2.725
|
NM_001042610
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr5_+_148651443
|
2.715
|
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr12_-_96184510
|
2.712
|
NM_021229
|
NTN4
|
netrin 4
|
|
chrX_+_102632108
|
2.692
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr11_-_128391888
|
2.685
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr2_+_170590454
|
2.676
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chrX_+_43514157
|
2.672
|
|
MAOA
|
monoamine oxidase A
|
|
chr11_-_33722285
|
2.670
|
NM_001166692
|
C11orf91
|
chromosome 11 open reading frame 91
|
|
chr3_+_36421944
|
2.661
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr6_-_160114318
|
2.661
|
NM_000636
NM_001024465
NM_001024466
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr4_-_170191989
|
2.646
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr6_+_151561653
|
2.646
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_-_143059696
|
2.643
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr16_+_3096681
|
2.643
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr11_-_32452358
|
2.640
|
NM_001198551
NM_001198552
|
WT1
|
Wilms tumor 1
|
|
chr17_-_29151724
|
2.624
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chrY_-_1521733
|
2.608
|
NM_001173474
NM_004192
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr8_-_145578502
|
2.597
|
NM_001252402
NM_001252404
|
C8ORFK29
|
uncharacterized LOC340393
|