|
chr8_-_22550456
|
17.150
|
|
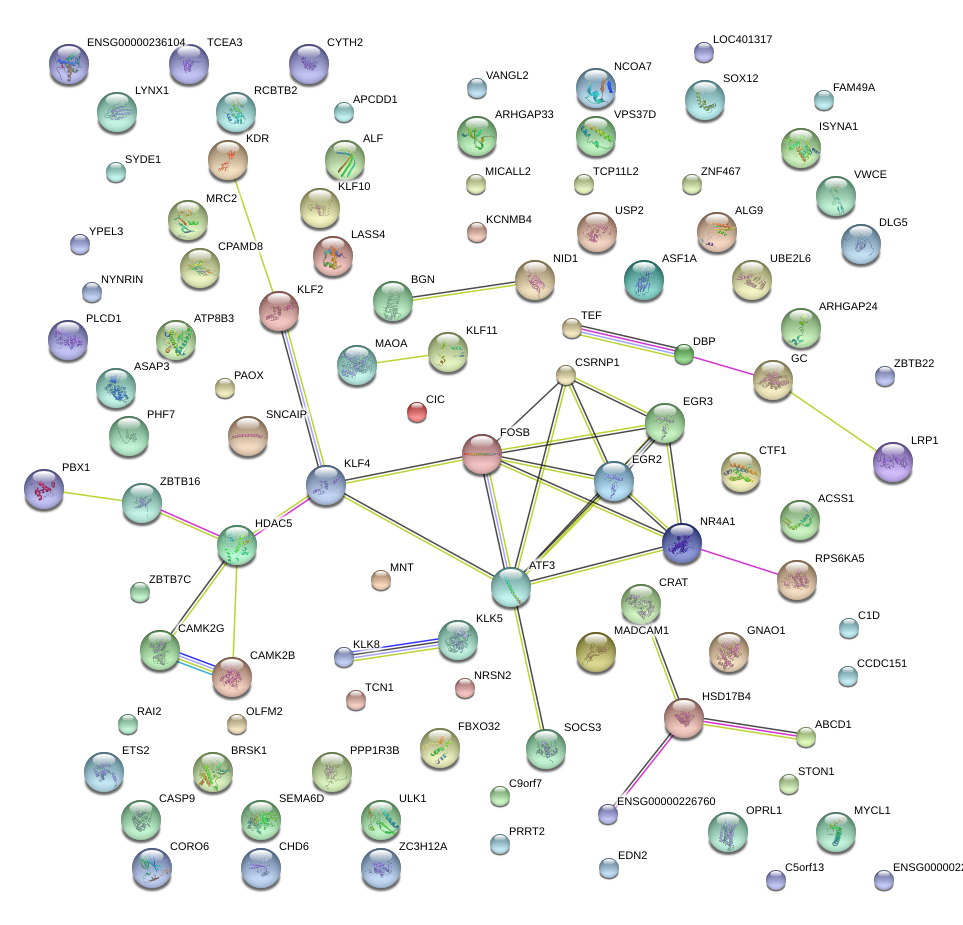
EGR3
|
early growth response 3
|
|
chr1_+_212781969
|
14.568
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_17007674
|
13.025
|
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr8_-_22550708
|
12.826
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_+_212782103
|
11.854
|
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_1812198
|
8.757
|
NM_001178002
NM_138813
|
ATP8B3
|
ATPase, aminophospholipid transporter, class I, type 8B, member 3
|
|
chr1_+_37940129
|
7.186
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr10_+_135192694
|
7.154
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr12_+_52445185
|
7.015
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr4_+_86396251
|
6.800
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_-_119234628
|
6.689
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_+_56225250
|
6.631
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr8_-_143858436
|
6.556
|
NM_177476
NM_177477
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr1_+_37940074
|
6.535
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr6_+_126111782
|
6.511
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr20_+_62711434
|
6.402
|
NM_000913
NM_182647
|
OPRL1
|
opiate receptor-like 1
|
|
chr9_-_131872907
|
6.257
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr1_+_37940180
|
6.148
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr6_+_126112057
|
5.968
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr12_+_52416615
|
5.799
|
NM_001202233
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr3_-_39195034
|
5.799
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr1_-_15850776
|
5.798
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr3_-_39195101
|
5.582
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr19_+_16435734
|
5.451
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr17_-_27949814
|
5.423
|
|
CORO6
|
coronin 6
|
|
chr19_+_8274179
|
5.344
|
NM_024552
|
CERS4
|
ceramide synthase 4
|
|
chr12_+_57522247
|
5.334
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr19_+_45971249
|
5.268
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr6_-_33285493
|
5.238
|
NM_005453
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr4_-_55991746
|
5.208
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr1_-_23810627
|
5.172
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr9_-_131872782
|
5.130
|
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr1_+_164528586
|
5.130
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr19_+_16435632
|
5.108
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_-_119234859
|
5.107
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr21_+_40177847
|
5.046
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr7_+_73082157
|
5.045
|
NM_001077621
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr18_-_45935627
|
4.937
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr17_+_60704747
|
4.884
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr1_-_40367539
|
4.687
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr8_-_124553253
|
4.655
|
|
FBXO32
|
F-box protein 32
|
|
chr9_-_110251754
|
4.626
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr2_-_16847070
|
4.516
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr19_+_36266366
|
4.515
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr16_-_30107492
|
4.493
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr16_+_29823408
|
4.470
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr3_-_38071030
|
4.419
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr18_+_10454620
|
4.347
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr8_-_143859177
|
4.320
|
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr19_-_32896441
|
4.319
|
|
LOC400684
|
uncharacterized LOC400684
|
|
chr19_+_15218141
|
4.314
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr8_-_103666018
|
4.314
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chrX_+_152990322
|
4.306
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr9_-_110252045
|
4.300
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_236228670
|
4.294
|
|
NID1
|
nidogen 1
|
|
chr1_-_23810721
|
4.271
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr14_+_24867926
|
4.270
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr5_+_121647773
|
4.261
|
NM_001242935
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr10_-_75634219
|
4.234
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr11_-_57335080
|
4.223
|
NM_004223
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr17_-_79008372
|
4.197
|
|
FLJ90757
|
uncharacterized LOC440465
|
|
chr3_+_52444514
|
4.193
|
NM_016483
NM_173341
|
PHF7
|
PHD finger protein 7
|
|
chr19_-_49140569
|
4.166
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr10_-_64576074
|
4.161
|
|
EGR2
|
early growth response 2
|
|
chr12_+_132379262
|
4.154
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr10_-_64576123
|
4.125
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr20_-_40246991
|
4.090
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr19_+_42788605
|
4.088
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr1_+_160370363
|
4.047
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr21_+_40177754
|
4.029
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr11_+_113930430
|
4.028
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr12_+_70759974
|
4.028
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr14_-_91526912
|
4.020
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr17_-_76356153
|
4.008
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr9_+_136325086
|
3.986
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr20_+_327725
|
3.969
|
|
NRSN2
|
neurensin 2
|
|
chr19_+_496453
|
3.968
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr4_-_174090305
|
3.962
|
|
|
|
|
chr6_-_33285718
|
3.950
|
NM_001145338
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr17_-_42201009
|
3.949
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr19_-_7701852
|
3.947
|
|
|
|
|
chr19_-_51504799
|
3.929
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr20_-_25038501
|
3.918
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr12_+_57522429
|
3.895
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr22_+_41777932
|
3.893
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr15_+_48010685
|
3.888
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr7_+_73082320
|
3.885
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr17_-_42200957
|
3.883
|
|
HDAC5
|
histone deacetylase 5
|
|
chr17_-_2304217
|
3.838
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr19_-_18548788
|
3.823
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chrX_+_152760346
|
3.814
|
NM_001711
|
BGN
|
biglycan
|
|
chr1_-_23751126
|
3.756
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr19_-_10047014
|
3.746
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr19_+_55795766
|
3.641
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr13_-_49107185
|
3.636
|
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr11_+_113930296
|
3.636
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr16_+_30907927
|
3.603
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr1_-_41950296
|
3.597
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chrX_-_17879355
|
3.588
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr19_+_48972464
|
3.573
|
NM_004228
NM_017457
|
CYTH2
|
cytohesin 2
|
|
chrX_+_43514157
|
3.570
|
|
MAOA
|
monoamine oxidase A
|
|
chr5_-_111093251
|
3.559
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_+_106696508
|
3.555
|
NM_152772
|
TCP11L2
|
t-complex 11 (mouse)-like 2
|
|
chr2_+_10183630
|
3.547
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr7_-_149470294
|
3.525
|
NM_207336
|
ZNF467
|
zinc finger protein 467
|
|
chr10_-_79686279
|
3.524
|
NM_004747
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr5_+_121647385
|
3.494
|
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr11_-_61062534
|
3.482
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr8_+_96281040
|
3.479
|
|
LOC100616530
|
tospeak
|
|
chr8_-_9008138
|
3.479
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr7_-_1498961
|
3.465
|
|
MICALL2
|
MICAL-like 2
|
|
chr2_+_48757292
|
3.459
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr19_-_51456159
|
3.441
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr16_+_56225301
|
3.437
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr20_+_306205
|
3.420
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr16_+_29823557
|
3.410
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr10_-_98592186
|
3.398
|
|
|
|
|
chr7_+_28448970
|
3.381
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr6_+_19837599
|
3.378
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chrX_+_134555867
|
3.356
|
|
LINC00086
|
long intergenic non-protein coding RNA 86
|
|
chr19_+_39897457
|
3.294
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr19_-_36523770
|
3.288
|
NM_015526
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr8_-_124553448
|
3.274
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr3_-_187871816
|
3.264
|
|
LPP-AS2
|
LPP antisense RNA 2 (non-protein coding)
|
|
chr17_+_60705051
|
3.262
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr13_+_88324777
|
3.248
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr15_+_63569748
|
3.231
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr15_+_65134081
|
3.197
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chrX_-_53350521
|
3.189
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr13_+_52027435
|
3.167
|
|
|
|
|
chr8_-_96281399
|
3.154
|
NM_177965
|
C8orf37
|
chromosome 8 open reading frame 37
|
|
chr7_+_2559396
|
3.153
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr11_+_5617330
|
3.151
|
NM_058166
|
TRIM6
|
tripartite motif containing 6
|
|
chr19_-_8455552
|
3.140
|
|
LOC100507567
|
uncharacterized LOC100507567
|
|
chr7_-_102257199
|
3.136
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr7_-_1499107
|
3.133
|
NM_182924
|
MICALL2
|
MICAL-like 2
|
|
chr2_+_30369868
|
3.133
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr17_+_60705042
|
3.132
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr19_+_48972574
|
3.131
|
|
CYTH2
|
cytohesin 2
|
|
chr19_-_18548118
|
3.122
|
NM_001253389
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr2_+_48757063
|
3.102
|
NM_001198593
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr7_+_73868236
|
3.102
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr18_+_10454542
|
3.101
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr1_-_115632047
|
3.099
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr1_+_164528914
|
3.092
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chrX_+_102632108
|
3.090
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr17_-_33775761
|
3.082
|
NM_144682
|
SLFN13
|
schlafen family member 13
|
|
chr3_+_52444673
|
3.075
|
|
PHF7
|
PHD finger protein 7
|
|
chr14_+_75745480
|
3.068
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr18_+_3449972
|
3.065
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr16_-_1993189
|
3.051
|
|
SEPX1
|
selenoprotein X, 1
|
|
chr13_-_110438896
|
3.045
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr6_+_44095357
|
3.044
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr22_-_30685485
|
3.040
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr5_-_134914968
|
3.024
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr2_-_96811138
|
3.012
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr16_-_325867
|
2.997
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr11_-_94964209
|
2.993
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr5_-_139725860
|
2.991
|
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr11_-_46142955
|
2.991
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr1_+_3569098
|
2.976
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr7_+_73868397
|
2.974
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr2_+_30369800
|
2.972
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr6_-_41863086
|
2.969
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr7_-_82792127
|
2.964
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr1_+_43282775
|
2.962
|
NM_001017922
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chrX_-_43741593
|
2.931
|
NM_000898
|
MAOB
|
monoamine oxidase B
|
|
chr2_+_20646831
|
2.912
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr11_+_62380212
|
2.911
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr3_-_105587710
|
2.907
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr3_-_38066277
|
2.897
|
NM_001130964
|
PLCD1
|
phospholipase C, delta 1
|
|
chr8_-_143484528
|
2.885
|
NM_145003
|
TSNARE1
|
t-SNARE domain containing 1
|
|
chr6_+_138188352
|
2.883
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr9_-_23821808
|
2.878
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr17_-_36762105
|
2.868
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr18_+_11981457
|
2.861
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr18_+_10454625
|
2.854
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr1_-_108507474
|
2.853
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr12_-_57824787
|
2.849
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr18_+_11981421
|
2.834
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr19_-_36523540
|
2.833
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr9_-_20622433
|
2.829
|
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr11_-_560706
|
2.824
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr5_+_172484353
|
2.818
|
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr11_+_118478305
|
2.794
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr19_-_51487070
|
2.791
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr17_-_27038871
|
2.785
|
NM_152465
|
PROCA1
|
protein interacting with cyclin A1
|
|
chr1_-_54871681
|
2.785
|
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr2_-_96810775
|
2.779
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr8_+_1711869
|
2.778
|
NM_018941
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr3_+_49208979
|
2.777
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr10_+_72432557
|
2.777
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr1_+_43282796
|
2.772
|
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chr11_-_1593149
|
2.771
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr10_+_23728197
|
2.769
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr12_-_133532847
|
2.768
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr20_-_56884488
|
2.759
|
|
PPP4R1L
|
protein phosphatase 4, regulatory subunit 1-like
|
|
chr3_-_139396773
|
2.754
|
NM_001200047
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr13_-_49107250
|
2.749
|
NM_001268
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr19_+_36359346
|
2.746
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|