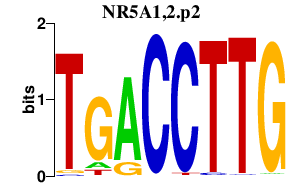
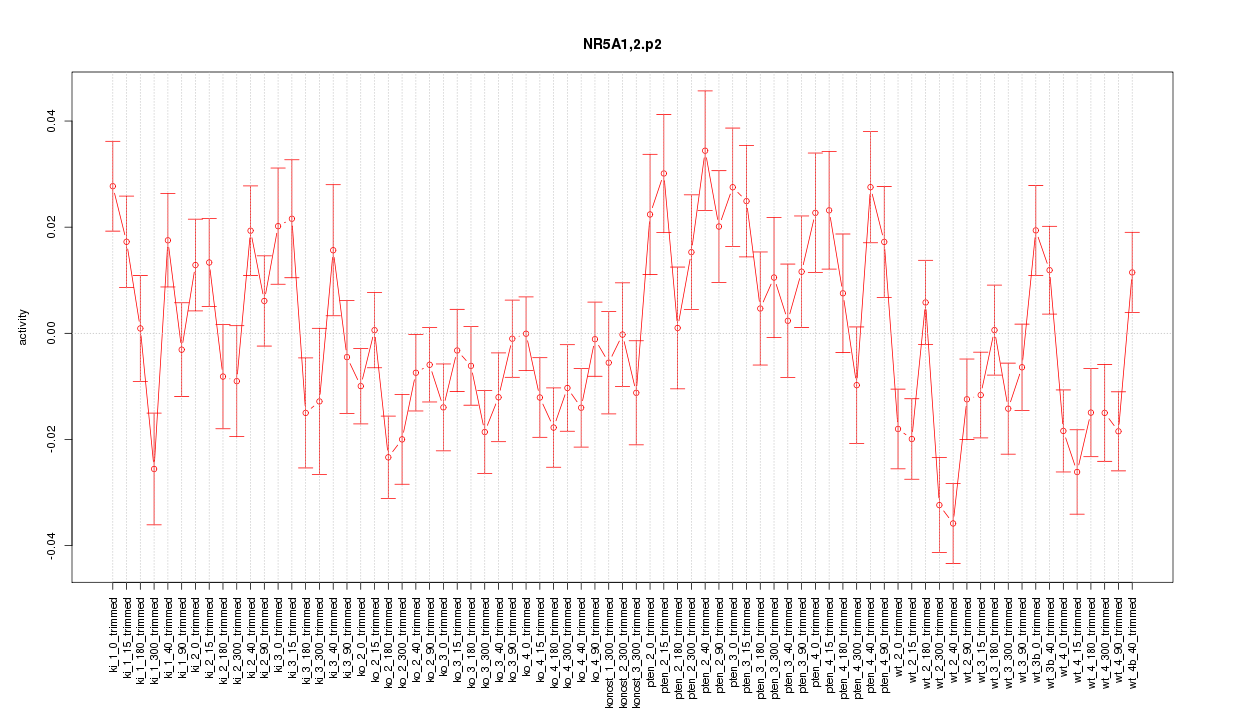
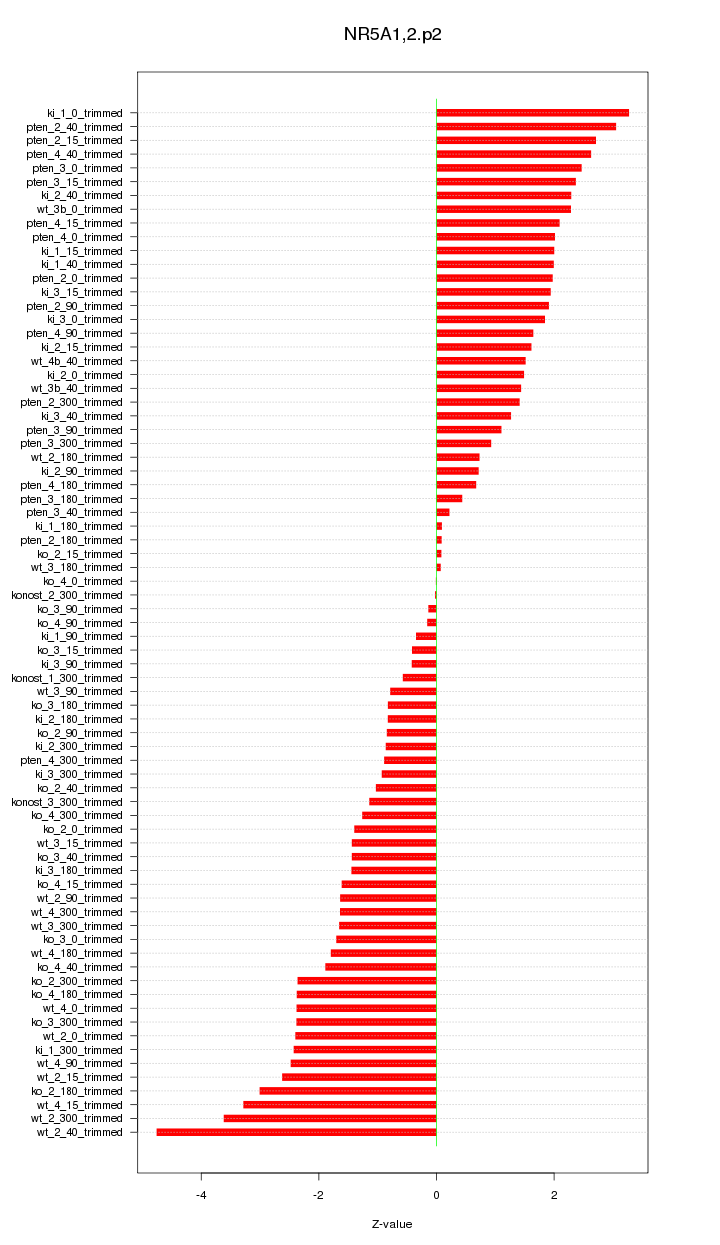
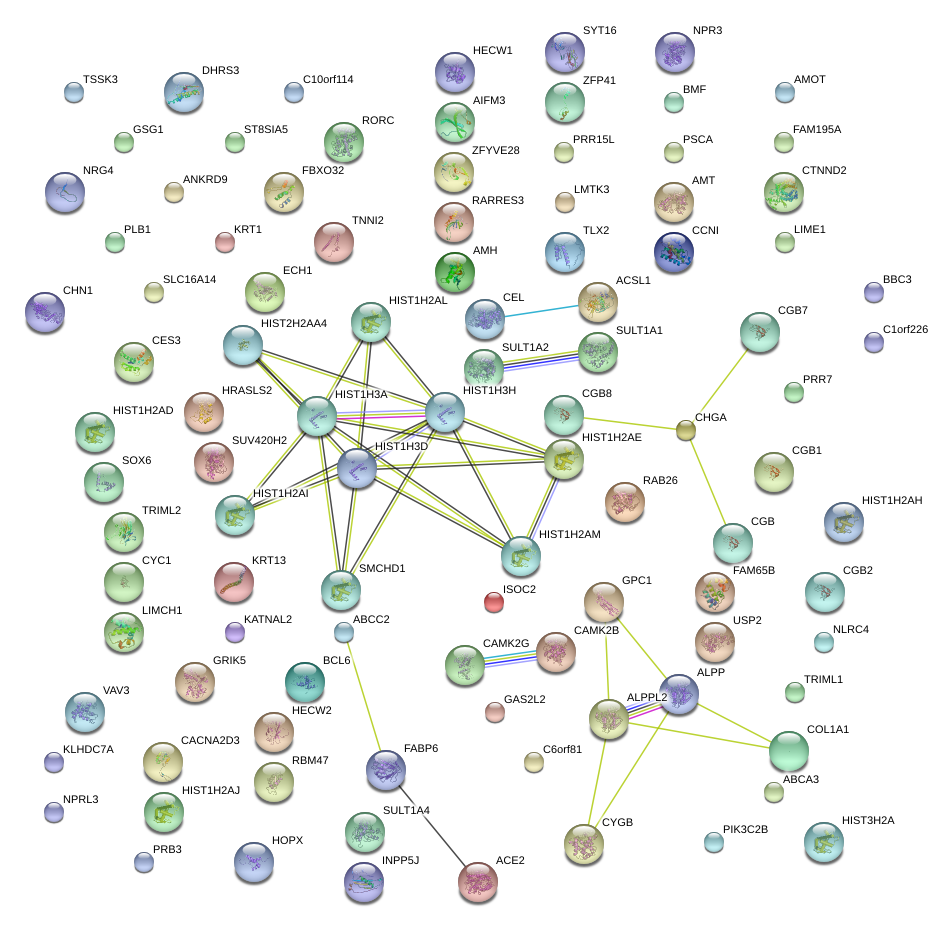
|
chr11_-_119234628
|
12.725
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_29309092
|
12.686
|
|
|
|
|
chr6_-_27782517
|
8.458
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr4_-_57522469
|
8.344
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr2_+_241375207
|
7.505
|
|
GPC1
|
glypican 1
|
|
chr4_-_189026401
|
7.221
|
NM_173553
|
TRIML2
|
tripartite motif family-like 2
|
|
chr15_-_76352068
|
7.008
|
|
NRG4
|
neuregulin 4
|
|
chr7_+_43152196
|
6.899
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr10_+_101542460
|
6.679
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr22_+_31518923
|
6.625
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr6_-_26199439
|
6.553
|
NM_021065
NM_003530
|
HIST1H2AD
HIST1H3D
|
histone cluster 1, H2ad
histone cluster 1, H3d
|
|
chr2_-_175869910
|
6.485
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr5_-_11903964
|
6.483
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr17_-_48266572
|
6.482
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr17_-_48266313
|
6.476
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_32827843
|
6.432
|
NM_001167676
|
LOC100128071
|
uncharacterized LOC100128071
|
|
chr11_+_1860718
|
6.418
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr19_+_2249112
|
6.395
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr12_-_123201319
|
6.256
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr4_-_57522059
|
6.215
|
|
HOPX
|
HOP homeobox
|
|
chrX_-_130964633
|
6.162
|
|
LOC286467
|
family with sequence similarity 195, member A pseudogene
|
|
chr22_+_21321446
|
6.021
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr10_+_101542554
|
5.737
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr12_-_123187842
|
5.732
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr2_-_230933618
|
5.658
|
NM_152527
|
SLC16A14
|
solute carrier family 16, member 14 (monocarboxylic acid transporter 14)
|
|
chr4_+_189060597
|
5.629
|
NM_178556
|
TRIML1
|
tripartite motif family-like 1
|
|
chr17_-_46035109
|
5.558
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr10_-_21786186
|
5.502
|
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr4_+_41614911
|
5.431
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_-_21786205
|
5.410
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr17_-_48271968
|
5.401
|
|
|
|
|
chr1_-_151798552
|
5.233
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr1_+_18807423
|
5.200
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr4_+_41614933
|
5.124
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr19_+_55851220
|
5.116
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr11_-_119234859
|
5.073
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_+_2198607
|
5.056
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr12_-_53074172
|
5.053
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr12_-_13248581
|
5.011
|
NM_001206843
NM_001206845
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr8_+_143761873
|
4.879
|
NM_005672
|
PSCA
|
prostate stem cell antigen
|
|
chr17_-_39661803
|
4.845
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr11_-_63330854
|
4.826
|
NM_017878
|
HRASLS2
|
HRAS-like suppressor 2
|
|
chr19_-_49558996
|
4.653
|
NM_033142
|
CGB7
|
chorionic gonadotropin, beta polypeptide 7
|
|
chr4_+_41614725
|
4.637
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_-_49460110
|
4.496
|
NM_000481
NM_001164710
NM_001164711
NM_001164712
|
AMT
|
aminomethyltransferase
|
|
chr19_-_42569921
|
4.494
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr3_-_187454247
|
4.363
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_-_49016418
|
4.310
|
NM_001080434
|
LMTK3
|
lemur tyrosine kinase 3
|
|
chr12_-_11422640
|
4.296
|
NM_006249
|
PRB3
|
proline-rich protein BstNI subfamily 3
|
|
chrX_-_15620191
|
4.294
|
NM_021804
|
ACE2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr6_-_24877453
|
4.266
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr16_-_67223998
|
4.263
|
NM_178516
|
EXOC3L1
|
exocyst complex component 3-like 1
|
|
chr11_+_61248584
|
4.257
|
NM_001170753
NM_145017
|
PPP1R32
|
protein phosphatase 1, regulatory subunit 32
|
|
chr17_-_74533623
|
4.141
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr4_-_185726865
|
4.114
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr2_+_28718937
|
4.046
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr1_+_32827797
|
3.983
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr1_-_12677347
|
3.975
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr2_+_233243243
|
3.975
|
NM_001632
|
ALPP
|
alkaline phosphatase, placental
|
|
chr1_-_204459445
|
3.952
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr17_-_48265912
|
3.951
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr16_+_2198687
|
3.943
|
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr2_-_32490688
|
3.896
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr19_-_55973048
|
3.884
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr6_+_27833033
|
3.856
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr16_-_28608367
|
3.854
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr8_+_145149985
|
3.816
|
|
CYC1
|
cytochrome c-1
|
|
chr5_+_32710742
|
3.767
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr17_-_34079896
|
3.738
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr14_+_93389491
|
3.691
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr3_+_54156691
|
3.654
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr9_+_135937364
|
3.643
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr1_+_9415944
|
3.613
|
|
|
|
|
chr18_+_44497512
|
3.591
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr5_+_176875325
|
3.577
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr5_+_159656436
|
3.575
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr8_-_124553253
|
3.529
|
|
FBXO32
|
F-box protein 32
|
|
chr4_-_2420252
|
3.521
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chrX_-_112084006
|
3.514
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr15_-_40398286
|
3.457
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr10_-_75634219
|
3.449
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr16_+_691901
|
3.403
|
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr22_+_22676814
|
3.360
|
|
|
|
|
chr11_-_16424391
|
3.309
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr14_+_62462540
|
3.303
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr19_-_47735858
|
3.258
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr11_+_63304272
|
3.253
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr5_-_11904127
|
3.220
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr16_+_67001377
|
3.217
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr1_+_162351519
|
3.215
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr19_+_50304801
|
3.183
|
|
|
|
|
chr2_+_74741565
|
3.171
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr6_+_35704858
|
3.168
|
NM_145028
|
C6orf81
|
chromosome 6 open reading frame 81
|
|
chr19_-_39322411
|
3.150
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr14_-_102975996
|
3.074
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr8_+_144349564
|
3.062
|
NM_138465
|
GLI4
|
GLI family zinc finger 4
|
|
chr18_-_44336991
|
3.055
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr4_-_40517913
|
3.016
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_108507474
|
3.014
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr17_-_36762105
|
3.011
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr3_+_54156573
|
3.001
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr22_+_41865112
|
2.998
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr1_+_212738675
|
2.986
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr19_-_17356022
|
2.980
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr12_+_57849086
|
2.972
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr8_-_145752405
|
2.962
|
NM_001024678
|
LRRC24
|
leucine rich repeat containing 24
|
|
chr2_+_219646704
|
2.949
|
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr11_+_65658101
|
2.949
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr16_+_57481336
|
2.930
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr19_-_47734215
|
2.927
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr8_+_145149978
|
2.922
|
|
CYC1
|
cytochrome c-1
|
|
chr2_+_98262497
|
2.913
|
NM_001862
|
COX5B
|
cytochrome c oxidase subunit Vb
|
|
chr3_+_160394946
|
2.876
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr22_+_50354428
|
2.864
|
|
PIM3
|
pim-3 oncogene
|
|
chr10_+_99344421
|
2.861
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr3_-_49459870
|
2.795
|
|
AMT
|
aminomethyltransferase
|
|
chr4_-_149357954
|
2.790
|
|
|
|
|
chr12_+_13154435
|
2.787
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr19_+_2328679
|
2.783
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr7_-_27187367
|
2.779
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr6_-_27806116
|
2.762
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr17_-_45899133
|
2.760
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr13_+_31287614
|
2.746
|
NM_001204406
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr11_+_69061599
|
2.745
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr17_-_48271365
|
2.744
|
|
|
|
|
chr10_+_70847857
|
2.738
|
|
SRGN
|
serglycin
|
|
chr9_+_37650996
|
2.732
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr17_+_48545689
|
2.722
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr10_-_101190201
|
2.710
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr16_-_66952729
|
2.687
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr6_+_152128453
|
2.684
|
NM_001122740
|
ESR1
|
estrogen receptor 1
|
|
chr19_+_54965628
|
2.683
|
|
|
|
|
chr7_+_94536926
|
2.674
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr8_+_145149936
|
2.671
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr7_-_64451413
|
2.665
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr7_+_150688143
|
2.658
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr12_+_13153337
|
2.640
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr3_-_187871816
|
2.637
|
|
LPP-AS2
|
LPP antisense RNA 2 (non-protein coding)
|
|
chr22_+_18043147
|
2.626
|
NM_031481
|
SLC25A18
|
solute carrier family 25 (mitochondrial carrier), member 18
|
|
chr15_+_63050761
|
2.609
|
|
TLN2
|
talin 2
|
|
chr17_-_37381920
|
2.588
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr19_+_2328628
|
2.570
|
NM_001077238
NM_152988
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chrX_-_112066353
|
2.567
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr17_-_73505914
|
2.564
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr7_+_128312345
|
2.559
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr2_-_202222066
|
2.555
|
NM_001127391
NM_139163
|
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12
|
|
chr17_-_48546174
|
2.544
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr10_+_26727651
|
2.536
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr6_+_26199712
|
2.530
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr6_+_152128685
|
2.524
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr17_-_1553099
|
2.522
|
NM_031430
|
RILP
|
Rab interacting lysosomal protein
|
|
chr19_+_11350282
|
2.513
|
NM_018687
|
C19orf80
|
chromosome 19 open reading frame 80
|
|
chr16_-_28621279
|
2.505
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr16_-_4665002
|
2.492
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr1_-_26147256
|
2.489
|
|
LOC646471
|
uncharacterized LOC646471
|
|
chr1_+_15764934
|
2.488
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr16_+_776957
|
2.476
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr8_-_108510094
|
2.474
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr19_+_49468656
|
2.472
|
|
FTL
|
ferritin, light polypeptide
|
|
chr1_+_6094342
|
2.470
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr12_-_106532447
|
2.468
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chrX_-_65835845
|
2.465
|
NM_001242310
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr4_+_86749045
|
2.465
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr4_-_40631846
|
2.457
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr5_-_160279045
|
2.447
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr5_+_172068188
|
2.446
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr7_+_141478241
|
2.426
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chr17_+_48049678
|
2.401
|
|
DLX4
|
distal-less homeobox 4
|
|
chr19_+_2328685
|
2.385
|
|
SPPL2B
|
signal peptide peptidase-like 2B
|
|
chr17_-_45899118
|
2.374
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr6_-_41130921
|
2.362
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr11_-_8190535
|
2.353
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr1_-_59248287
|
2.348
|
|
JUN
|
jun proto-oncogene
|
|
chr5_+_145317297
|
2.347
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr13_-_114103452
|
2.342
|
NM_199162
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
|
chr6_-_11232883
|
2.334
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11232900
|
2.332
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr10_-_94050799
|
2.327
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr6_-_27860927
|
2.318
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr15_-_40401041
|
2.313
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr1_-_155178968
|
2.302
|
|
THBS3
|
thrombospondin 3
|
|
chr6_-_11232901
|
2.299
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_27775683
|
2.299
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr14_+_78266419
|
2.288
|
NM_001142545
NM_020421
|
ADCK1
|
aarF domain containing kinase 1
|
|
chr20_+_43803539
|
2.278
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr14_-_102976096
|
2.278
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr2_+_111878450
|
2.277
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr2_+_233320832
|
2.275
|
NM_001631
|
ALPI
|
alkaline phosphatase, intestinal
|
|
chr19_+_6372720
|
2.270
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr16_-_54319966
|
2.268
|
|
IRX3
|
iroquois homeobox 3
|
|
chr3_+_160473995
|
2.255
|
NM_139245
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L
|
|
chr22_+_37257004
|
2.247
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr19_+_6372805
|
2.244
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr22_+_32010152
|
2.237
|
|
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast)
|
|
chr2_-_192711651
|
2.234
|
|
SDPR
|
serum deprivation response
|
|
chr7_+_2443205
|
2.231
|
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12
|
|
chr10_-_76995685
|
2.225
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr17_-_39093671
|
2.224
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr3_-_73673978
|
2.221
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr6_-_30899926
|
2.217
|
NM_205854
|
SFTA2
|
surfactant associated 2
|