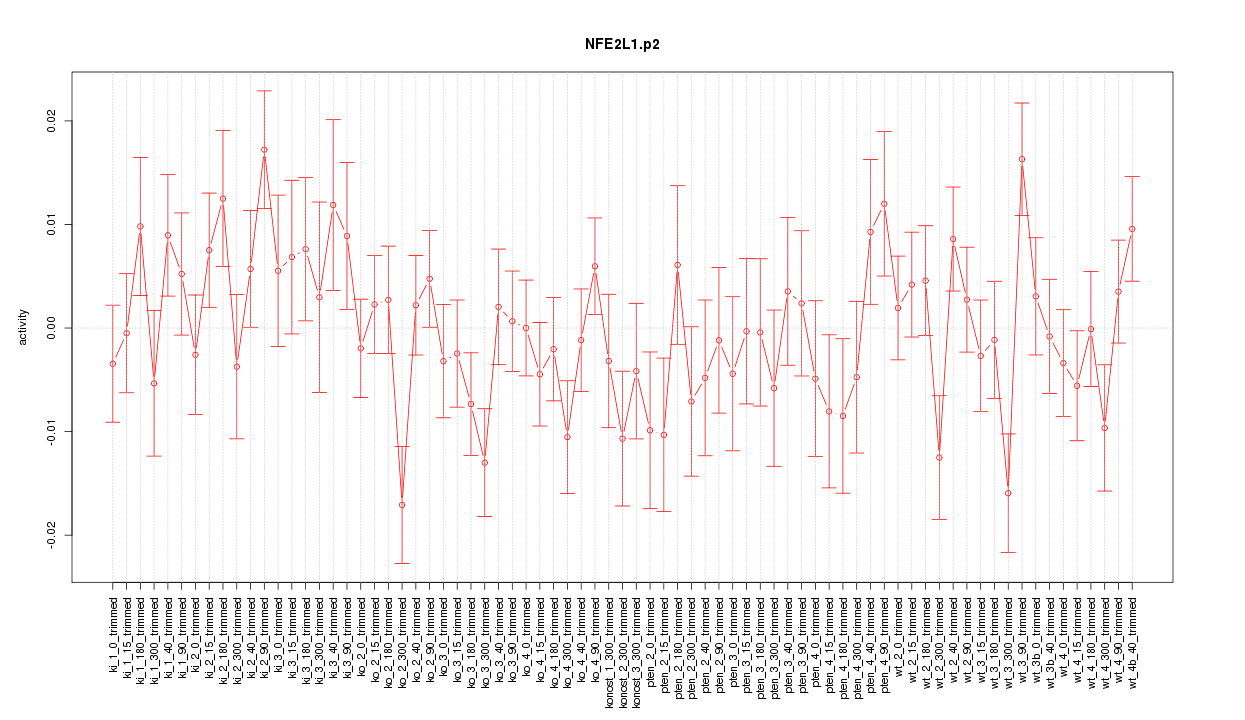
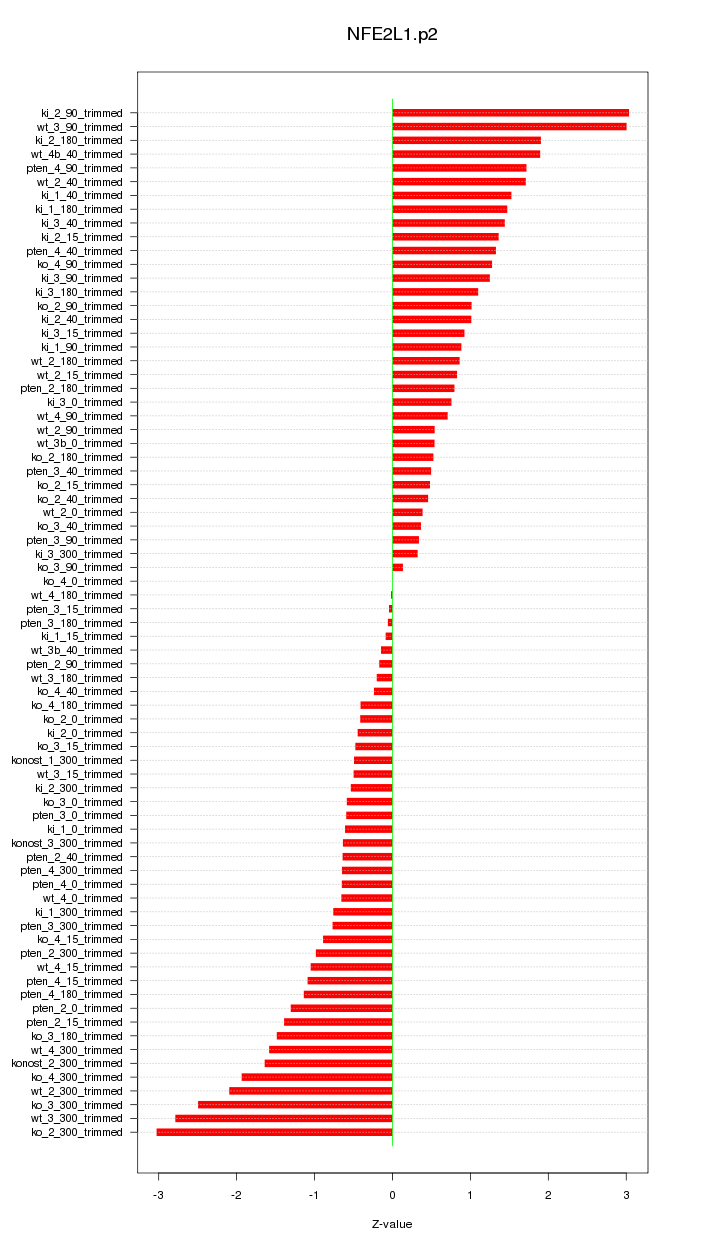
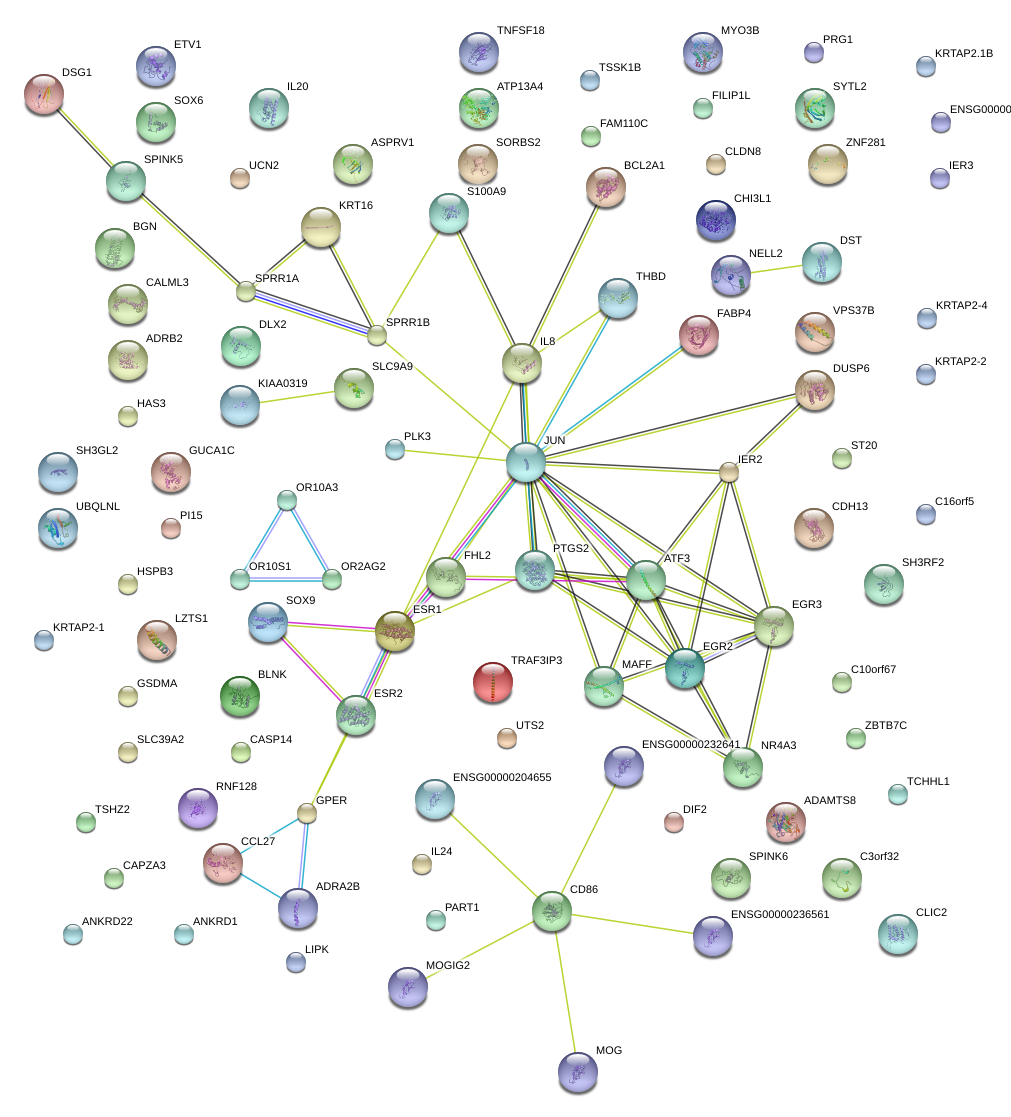
|
chr10_+_5566923
|
10.074
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr19_-_36019194
|
9.483
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr12_-_123187842
|
7.751
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr1_+_212738675
|
7.466
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_92680784
|
6.628
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr17_-_39216343
|
6.350
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr6_+_154897419
|
6.148
|
|
|
|
|
chr10_-_92681025
|
5.610
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr12_-_123201319
|
5.096
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr17_-_39222126
|
5.094
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr18_+_28898051
|
4.904
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr5_+_147582338
|
4.805
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr13_+_52034639
|
4.686
|
|
|
|
|
chr16_+_69139466
|
4.587
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_+_212781969
|
4.134
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr17_-_39211462
|
3.741
|
NM_033032
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr12_+_18891040
|
3.583
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr19_+_15160194
|
3.570
|
NM_012114
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase
|
|
chr6_-_56716412
|
3.523
|
|
DST
|
dystonin
|
|
chr9_+_102584136
|
3.495
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_+_207070787
|
3.475
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr5_+_145317297
|
3.369
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr11_-_85430117
|
3.352
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr4_+_74606222
|
3.343
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr21_-_31588242
|
3.254
|
NM_199328
|
CLDN8
|
claudin 8
|
|
chr3_-_143567163
|
3.135
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr5_+_59784004
|
3.124
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr11_-_16424391
|
3.103
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_-_80263460
|
3.049
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr2_+_113735595
|
3.017
|
NM_019618
|
IL36G
|
interleukin 36, gamma
|
|
chr2_-_70189396
|
2.938
|
NM_152792
|
ASPRV1
|
aspartic peptidase, retroviral-like 1
|
|
chr8_-_20112691
|
2.911
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr2_-_96781887
|
2.909
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr2_+_171034654
|
2.786
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr1_+_212782103
|
2.740
|
|
ATF3
|
activating transcription factor 3
|
|
chr17_+_38119225
|
2.723
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr1_-_203154304
|
2.697
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr20_-_23030141
|
2.644
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr10_-_64576074
|
2.605
|
|
EGR2
|
early growth response 2
|
|
chr1_-_59248287
|
2.584
|
|
JUN
|
jun proto-oncogene
|
|
chr10_-_64576123
|
2.567
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_98031154
|
2.527
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr18_-_45663667
|
2.510
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chrX_+_152770083
|
2.381
|
|
BGN
|
biglycan
|
|
chr6_+_152128685
|
2.375
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr19_+_13263890
|
2.374
|
|
IER2
|
immediate early response 2
|
|
chr11_-_6790187
|
2.315
|
NM_001004490
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2
|
|
chr2_-_106015423
|
2.306
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr16_+_82660390
|
2.281
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr8_-_82395401
|
2.277
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_+_45266048
|
2.273
|
|
PLK3
|
polo-like kinase 3
|
|
chr8_+_75736771
|
2.262
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr10_-_23633771
|
2.261
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr11_-_7961066
|
2.221
|
NM_001003745
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3
|
|
chr17_+_70117154
|
2.188
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr11_-_123848397
|
2.187
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr22_+_38597938
|
2.183
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr5_+_53751430
|
2.146
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr11_-_5537815
|
2.146
|
NM_145053
|
UBQLNL
|
ubiquilin-like
|
|
chrX_+_105969875
|
2.143
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr17_+_70117168
|
2.132
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr11_-_85430382
|
2.114
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr12_-_45307710
|
2.096
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_48601200
|
2.085
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr16_+_82660573
|
2.058
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr1_-_152061529
|
2.029
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr22_+_38598065
|
2.002
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr17_+_70117177
|
1.996
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr1_-_200377262
|
1.983
|
|
ZNF281
|
zinc finger protein 281
|
|
chr9_+_17578952
|
1.978
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr4_-_186578122
|
1.971
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_90484300
|
1.963
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr3_-_108672547
|
1.963
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr17_+_17398334
|
1.961
|
|
|
|
|
chr16_-_4562979
|
1.955
|
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr5_-_112770537
|
1.952
|
NM_032028
|
TSSK1B
|
testis-specific serine kinase 1B
|
|
chr1_-_29309092
|
1.942
|
|
|
|
|
chr3_+_121774208
|
1.942
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chrX_-_154563735
|
1.925
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr1_+_212788359
|
1.923
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr2_-_172967234
|
1.900
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr1_-_186649421
|
1.889
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr6_+_29624757
|
1.858
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr5_+_148206155
|
1.851
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr1_+_152956563
|
1.812
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr14_+_21467413
|
1.809
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr6_-_30712294
|
1.771
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr9_-_34662629
|
1.760
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr7_-_14025826
|
1.751
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr2_-_172967014
|
1.746
|
|
DLX2
|
distal-less homeobox 2
|
|
chr12_-_89746244
|
1.744
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr5_+_147443534
|
1.741
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr1_-_200377488
|
1.719
|
|
ZNF281
|
zinc finger protein 281
|
|
chr3_-_8693723
|
1.692
|
NM_015931
|
C3orf32
|
chromosome 3 open reading frame 32
|
|
chr2_-_46384
|
1.692
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr3_-_193272587
|
1.685
|
NM_032279
|
ATP13A4
|
ATPase type 13A4
|
|
chr1_+_153330329
|
1.675
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr12_-_123380682
|
1.656
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr4_-_186577858
|
1.650
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr20_+_51588945
|
1.649
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr17_-_39768939
|
1.647
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr1_+_209941894
|
1.645
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr8_-_22550708
|
1.633
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr2_-_172967627
|
1.615
|
|
DLX2
|
distal-less homeobox 2
|
|
chr3_-_99594915
|
1.615
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr10_-_90611675
|
1.602
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr7_+_1126442
|
1.587
|
NM_001039966
NM_001505
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr1_-_173020011
|
1.584
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr1_+_207039153
|
1.570
|
NM_018724
|
IL20
|
interleukin 20
|
|
chr11_+_124120422
|
1.562
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chrX_-_10535476
|
1.561
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_-_39203423
|
1.558
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr5_-_95748688
|
1.556
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr11_+_117947726
|
1.546
|
NM_001083947
NM_001173551
NM_001173552
NM_019894
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr1_+_161676761
|
1.534
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr10_+_5488521
|
1.529
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr16_+_86544233
|
1.525
|
|
FOXF1
|
forkhead box F1
|
|
chr18_-_56296188
|
1.509
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr11_+_76813884
|
1.499
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr11_-_57335080
|
1.487
|
NM_004223
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr12_-_89746238
|
1.487
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr11_+_117947785
|
1.484
|
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr1_-_146696896
|
1.476
|
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr12_-_89745997
|
1.470
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr7_+_129932973
|
1.462
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr11_-_6817013
|
1.454
|
NM_003696
|
OR6A2
|
olfactory receptor, family 6, subfamily A, member 2
|
|
chr1_+_100111430
|
1.452
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr22_+_38201300
|
1.436
|
|
H1F0
|
H1 histone family, member 0
|
|
chr3_-_108248168
|
1.435
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr7_+_29234325
|
1.419
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr16_+_82660641
|
1.418
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr11_-_26593667
|
1.394
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr2_-_161056573
|
1.388
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chrX_+_150345055
|
1.383
|
NM_004224
|
GPR50
|
G protein-coupled receptor 50
|
|
chr20_+_54823787
|
1.376
|
NM_019888
|
MC3R
|
melanocortin 3 receptor
|
|
chrX_+_70443055
|
1.376
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr2_-_165811995
|
1.372
|
NM_001199148
NM_173512
|
SLC38A11
|
solute carrier family 38, member 11
|
|
chr22_-_33402648
|
1.368
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr19_+_42259397
|
1.366
|
NM_002483
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen)
|
|
chr5_+_140613937
|
1.354
|
|
PCDHB18
|
protocadherin beta 18 pseudogene
|
|
chr7_+_107110501
|
1.350
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr19_-_7968426
|
1.341
|
|
|
|
|
chr5_+_36608430
|
1.340
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr17_-_3461288
|
1.332
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr7_-_82791799
|
1.331
|
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr7_-_82792127
|
1.330
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr1_+_45265896
|
1.330
|
|
PLK3
|
polo-like kinase 3
|
|
chr4_+_3465032
|
1.321
|
NM_001164673
NM_173660
|
DOK7
|
docking protein 7
|
|
chr17_-_46671043
|
1.289
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr16_-_4588738
|
1.287
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr7_+_80275967
|
1.284
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_+_164528914
|
1.270
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_-_154178783
|
1.265
|
NM_001010979
|
C1orf189
|
chromosome 1 open reading frame 189
|
|
chr12_+_109577201
|
1.264
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr12_-_8815370
|
1.258
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr22_+_38201232
|
1.248
|
|
H1F0
|
H1 histone family, member 0
|
|
chr6_+_55039070
|
1.248
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr3_-_45267068
|
1.245
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr17_-_39675078
|
1.230
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr19_-_51512803
|
1.227
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr12_-_123380608
|
1.226
|
|
|
|
|
chr12_-_48119224
|
1.226
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr12_-_123380628
|
1.214
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chrX_-_110513750
|
1.206
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr4_-_23891608
|
1.202
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr17_-_42081836
|
1.201
|
NM_004160
|
PYY
|
peptide YY
|
|
chr6_-_166401526
|
1.197
|
|
LINC00473
|
long intergenic non-protein coding RNA 473
|
|
chr12_-_57634474
|
1.195
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr20_+_327329
|
1.194
|
NM_024958
|
NRSN2
|
neurensin 2
|
|
chr6_+_72922513
|
1.191
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_-_105412466
|
1.190
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr5_-_11903964
|
1.183
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr3_+_53529030
|
1.180
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr17_+_57915192
|
1.175
|
|
|
|
|
chr17_+_4802866
|
1.175
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr15_-_74726258
|
1.172
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr10_-_5541401
|
1.171
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr12_-_10324675
|
1.171
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr2_-_231789940
|
1.169
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr11_-_88781039
|
1.165
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr14_+_23511375
|
1.162
|
NM_001099780
|
PSMB11
|
proteasome (prosome, macropain) subunit, beta type, 11
|
|
chr4_+_75230856
|
1.160
|
NM_001432
|
EREG
|
epiregulin
|
|
chr5_+_140579182
|
1.154
|
NM_018931
|
PCDHB11
|
protocadherin beta 11
|
|
chr12_+_54332575
|
1.153
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr19_-_9325546
|
1.139
|
NM_001005191
|
OR7D4
|
olfactory receptor, family 7, subfamily D, member 4
|
|
chr5_-_150948504
|
1.133
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr19_-_51522953
|
1.130
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr11_+_66188474
|
1.128
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr7_+_39017608
|
1.128
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr12_-_8815266
|
1.127
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr6_-_11779279
|
1.121
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr16_+_30759619
|
1.119
|
NM_000294
NM_001172432
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis)
|
|
chr4_+_184827882
|
1.118
|
|
|
|
|
chr19_-_10047014
|
1.114
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr11_+_6806247
|
1.112
|
NM_001004489
|
OR2AG1
|
olfactory receptor, family 2, subfamily AG, member 1
|
|
chr14_-_94423766
|
1.110
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr22_+_38201113
|
1.101
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr11_+_124134699
|
1.100
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr2_-_136873740
|
1.095
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr13_-_36429798
|
1.087
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|