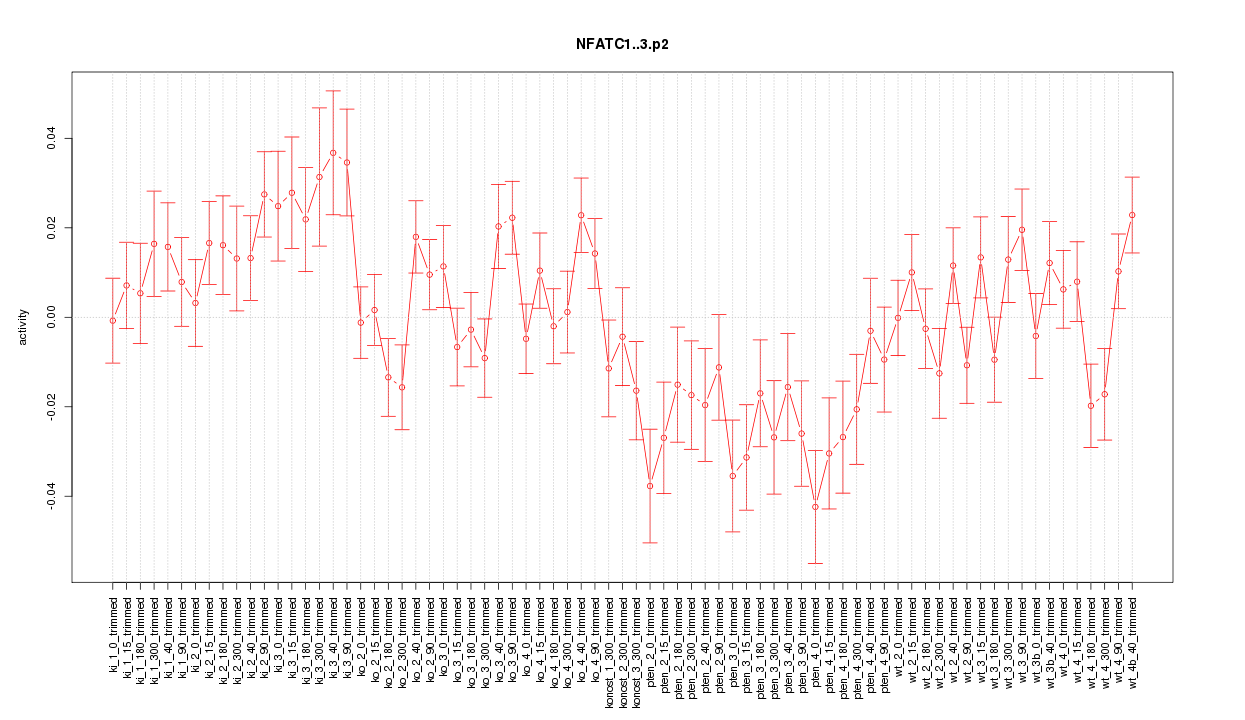
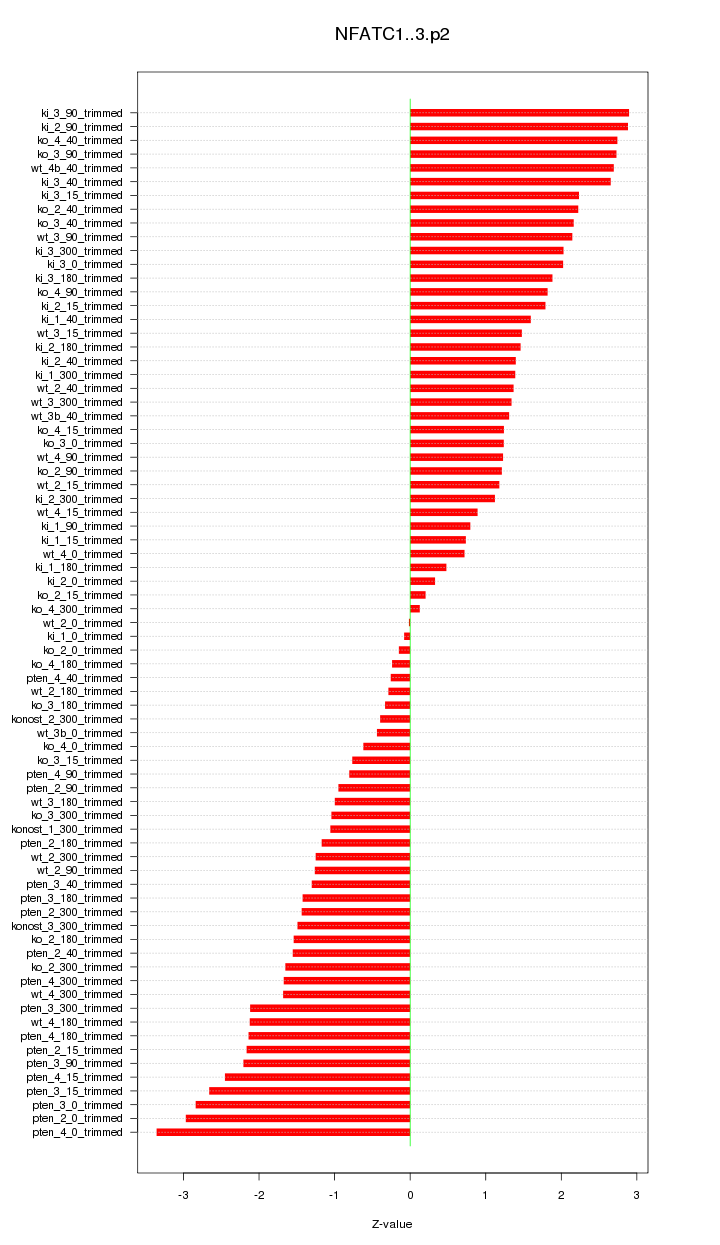
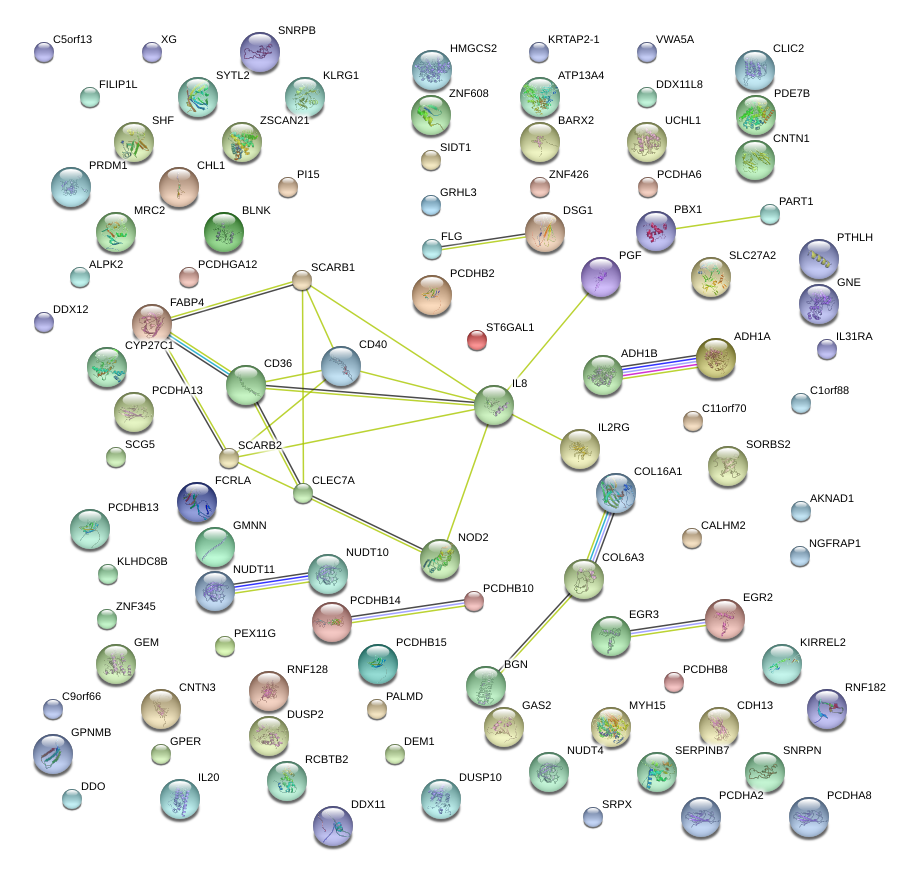
|
chr15_+_25101697
|
42.165
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr12_+_41086189
|
17.221
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr4_+_41258941
|
17.150
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr2_-_127963342
|
16.824
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr20_+_44746889
|
15.561
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr2_+_113735595
|
14.017
|
NM_019618
|
IL36G
|
interleukin 36, gamma
|
|
chr15_+_25230199
|
13.166
|
|
PAR5
|
Prader-Willi/Angelman syndrome-5
|
|
chr2_-_238322770
|
12.130
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322840
|
11.975
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr4_+_41258909
|
11.679
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr9_+_131870105
|
11.561
|
|
|
|
|
chr11_-_105010460
|
11.173
|
NM_021571
|
CARD18
|
caspase recruitment domain family, member 18
|
|
chr16_+_82660390
|
11.132
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr16_+_82660641
|
10.964
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr12_-_28124915
|
10.827
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr5_+_140602914
|
10.813
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chrX_-_70331297
|
10.807
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr6_+_106546736
|
10.758
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_+_140254930
|
10.628
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr16_+_82660573
|
10.020
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr15_+_25068793
|
9.916
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr8_-_82395401
|
9.521
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr2_-_238322806
|
9.095
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr8_-_22550057
|
9.051
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr4_+_41258895
|
8.804
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr5_+_140474198
|
8.681
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr18_+_28898051
|
8.620
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr1_+_161676761
|
8.452
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr7_+_1126442
|
8.370
|
NM_001039966
NM_001505
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr5_+_59784004
|
8.246
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr13_-_49110117
|
8.009
|
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr2_-_238322814
|
7.923
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrX_+_102632108
|
7.852
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr1_-_120311466
|
7.683
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr1_+_24645914
|
7.682
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr9_-_215892
|
7.458
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr2_-_238322815
|
7.356
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrX_+_152760346
|
7.302
|
NM_001711
|
BGN
|
biglycan
|
|
chr5_+_140593508
|
7.179
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr7_+_1127722
|
7.156
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr1_+_100111430
|
7.055
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr10_-_105212149
|
7.053
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr1_+_24645811
|
7.012
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr5_+_140566655
|
6.943
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr14_-_75422466
|
6.711
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr17_+_60704747
|
6.562
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr3_-_74570262
|
6.399
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chrX_-_38079980
|
6.396
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr5_+_140247767
|
6.245
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr17_-_39216343
|
6.222
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr3_+_49208979
|
6.180
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr20_+_58713521
|
6.146
|
|
LOC284757
|
uncharacterized LOC284757
|
|
chr15_+_50474357
|
6.124
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr15_+_25227140
|
5.957
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr2_-_96811138
|
5.878
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_+_40974430
|
5.822
|
NM_022774
|
DEM1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr10_-_105212022
|
5.705
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr19_+_36347809
|
5.687
|
NM_032123
NM_199179
NM_199180
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr5_+_140625146
|
5.645
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chrX_+_152760452
|
5.551
|
|
BGN
|
biglycan
|
|
chr11_+_101918168
|
5.537
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr1_+_111889194
|
5.444
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr6_+_136172802
|
5.414
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr1_-_221910801
|
5.405
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chrX_+_102631267
|
5.338
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr15_+_32933869
|
5.234
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr5_+_55147206
|
5.191
|
NM_001242637
NM_001242638
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr18_+_61420276
|
5.180
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr8_+_75736771
|
5.131
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr3_-_108248168
|
5.112
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr1_-_183387404
|
5.102
|
|
|
|
|
chr11_+_129245834
|
5.101
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr5_-_111093792
|
5.096
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_99594915
|
5.054
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr6_+_106534210
|
5.053
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_+_140174443
|
4.953
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr11_+_123986103
|
4.904
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr18_-_56296188
|
4.892
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr12_+_9142136
|
4.874
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chrX_-_154563735
|
4.836
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr1_-_109399705
|
4.834
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr19_-_9649257
|
4.798
|
NM_024106
|
ZNF426
|
zinc finger protein 426
|
|
chr15_-_45480160
|
4.792
|
|
SHF
|
Src homology 2 domain containing F
|
|
chrX_+_2670092
|
4.776
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chrX_-_51239295
|
4.752
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr10_-_98031154
|
4.661
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr3_-_193272587
|
4.610
|
NM_032279
|
ATP13A4
|
ATPase type 13A4
|
|
chr5_+_140207562
|
4.607
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr16_+_50731049
|
4.607
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr7_+_23286401
|
4.589
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_32169555
|
4.581
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chrX_+_105937067
|
4.580
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr1_-_32169619
|
4.553
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr6_+_13924676
|
4.547
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr8_-_95274540
|
4.432
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_-_10282680
|
4.407
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr4_+_74606222
|
4.407
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr1_+_207039153
|
4.402
|
NM_018724
|
IL20
|
interleukin 20
|
|
chr5_+_140809957
|
4.395
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr4_-_100242487
|
4.395
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr19_+_37342552
|
4.383
|
NM_001242475
NM_001242476
|
ZNF345
|
zinc finger protein 345
|
|
chr19_-_7553867
|
4.370
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr3_+_186739643
|
4.352
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr9_-_36276930
|
4.243
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr11_-_85437457
|
4.241
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_+_140261792
|
4.236
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr10_-_64578926
|
4.223
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr4_-_186697065
|
4.210
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr20_-_1165116
|
4.183
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr1_+_164528586
|
4.148
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_-_124080773
|
4.111
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr6_-_110736748
|
4.108
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr11_+_22696381
|
4.106
|
|
GAS2
|
growth arrest-specific 2
|
|
chr7_+_80267782
|
4.099
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_+_113251217
|
4.095
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr3_+_361365
|
4.093
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr19_+_42259397
|
4.064
|
NM_002483
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen)
|
|
chr1_+_247579457
|
4.045
|
NM_001079821
NM_001127461
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr5_-_146460991
|
4.037
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr9_+_22447353
|
3.976
|
|
DMRTA1
|
DMRT-like family A1
|
|
chr2_+_170366211
|
3.964
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr4_-_100140330
|
3.928
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr5_+_55149149
|
3.912
|
NM_001242636
NM_001242639
|
IL31RA
|
interleukin 31 receptor A
|
|
chr5_+_147648374
|
3.895
|
NM_001040129
|
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative)
|
|
chr3_-_161089747
|
3.890
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr16_+_84853585
|
3.882
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr1_-_221915393
|
3.850
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr11_-_102714240
|
3.846
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr7_+_23286390
|
3.813
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_140165875
|
3.812
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr5_-_115910406
|
3.810
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr6_-_159466019
|
3.793
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr5_+_140186658
|
3.789
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr4_+_166794409
|
3.779
|
NM_001204760
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr7_+_23286384
|
3.777
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr9_-_123688352
|
3.758
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr20_+_51801821
|
3.750
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr6_-_2635297
|
3.743
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chrX_-_84634664
|
3.734
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr20_+_123230
|
3.730
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr12_-_10605928
|
3.705
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr2_+_113885137
|
3.694
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr12_-_54694717
|
3.692
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr5_-_150284503
|
3.667
|
NM_001172831
NM_001172832
NM_052860
|
ZNF300
|
zinc finger protein 300
|
|
chr3_-_121740968
|
3.643
|
NM_001199799
NM_001199800
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr19_-_37341168
|
3.612
|
NM_001242801
|
ZNF790
|
zinc finger protein 790
|
|
chr7_+_23286386
|
3.608
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chrX_-_84634742
|
3.608
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr19_-_51472003
|
3.608
|
NM_001012964
NM_001012965
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr2_+_69201704
|
3.596
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr4_-_186577858
|
3.590
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_150738232
|
3.563
|
|
CTSS
|
cathepsin S
|
|
chr21_-_27945267
|
3.554
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr11_+_27015833
|
3.551
|
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chrX_-_69269781
|
3.538
|
NM_001002254
|
AWAT2
|
acyl-CoA wax alcohol acyltransferase 2
|
|
chr10_-_52008312
|
3.525
|
NM_001143974
NM_019893
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2
|
|
chr2_+_113885145
|
3.522
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr1_+_152769226
|
3.521
|
NM_178352
|
LCE1D
|
late cornified envelope 1D
|
|
chr12_+_100750856
|
3.507
|
NM_001145288
NM_139319
|
SLC17A8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8
|
|
chr17_-_39675078
|
3.504
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr3_-_172241060
|
3.490
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chrX_+_107288199
|
3.487
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr12_+_9980076
|
3.485
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr1_-_221915426
|
3.453
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr6_+_39760772
|
3.450
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr2_+_159651828
|
3.447
|
NM_001017920
|
DAPL1
|
death associated protein-like 1
|
|
chr1_-_221915458
|
3.432
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chrX_+_102631408
|
3.414
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr2_-_219858120
|
3.374
|
NM_005209
NM_057093
NM_057094
|
CRYBA2
|
crystallin, beta A2
|
|
chr5_+_140557370
|
3.344
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr5_+_140514787
|
3.340
|
NM_015669
|
PCDHB5
|
protocadherin beta 5
|
|
chr5_+_145317297
|
3.309
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr11_+_18194383
|
3.309
|
NM_054032
|
MRGPRX4
|
MAS-related GPR, member X4
|
|
chr10_-_62703822
|
3.284
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr12_-_11244828
|
3.242
|
NM_176884
|
TAS2R43
TAS2R31
|
taste receptor, type 2, member 43
taste receptor, type 2, member 31
|
|
chr1_+_164818083
|
3.239
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_39195101
|
3.236
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr14_-_106573348
|
3.226
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_165811995
|
3.196
|
NM_001199148
NM_173512
|
SLC38A11
|
solute carrier family 38, member 11
|
|
chr1_-_183274008
|
3.185
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr11_-_68780667
|
3.164
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chrX_-_84634396
|
3.162
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr5_-_141993691
|
3.135
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_183387501
|
3.129
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr1_+_150266261
|
3.124
|
NM_018997
NM_031901
|
MRPS21
|
mitochondrial ribosomal protein S21
|
|
chr7_+_28452143
|
3.122
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_140480056
|
3.093
|
NM_018937
|
PCDHB3
|
protocadherin beta 3
|
|
chrX_-_114252132
|
3.088
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr11_+_15133969
|
3.078
|
NM_001031853
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr5_-_64777703
|
3.075
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr1_+_248524882
|
3.068
|
NM_001004696
|
OR2T4
|
olfactory receptor, family 2, subfamily T, member 4
|
|
chr1_-_116383323
|
3.061
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr8_-_95274520
|
3.051
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_-_123411185
|
3.043
|
|
MYLK
|
myosin light chain kinase
|
|
chr1_+_161228516
|
3.029
|
NM_001102566
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
|
chr10_+_63422718
|
3.012
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr7_-_112727832
|
3.006
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_+_68800003
|
3.006
|
NM_001205255
|
OCLN
|
occludin
|
|
chr18_-_28681885
|
3.003
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_-_160549226
|
2.990
|
|
CD84
|
CD84 molecule
|