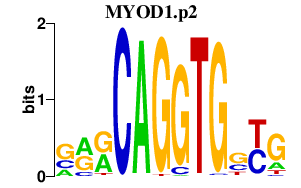
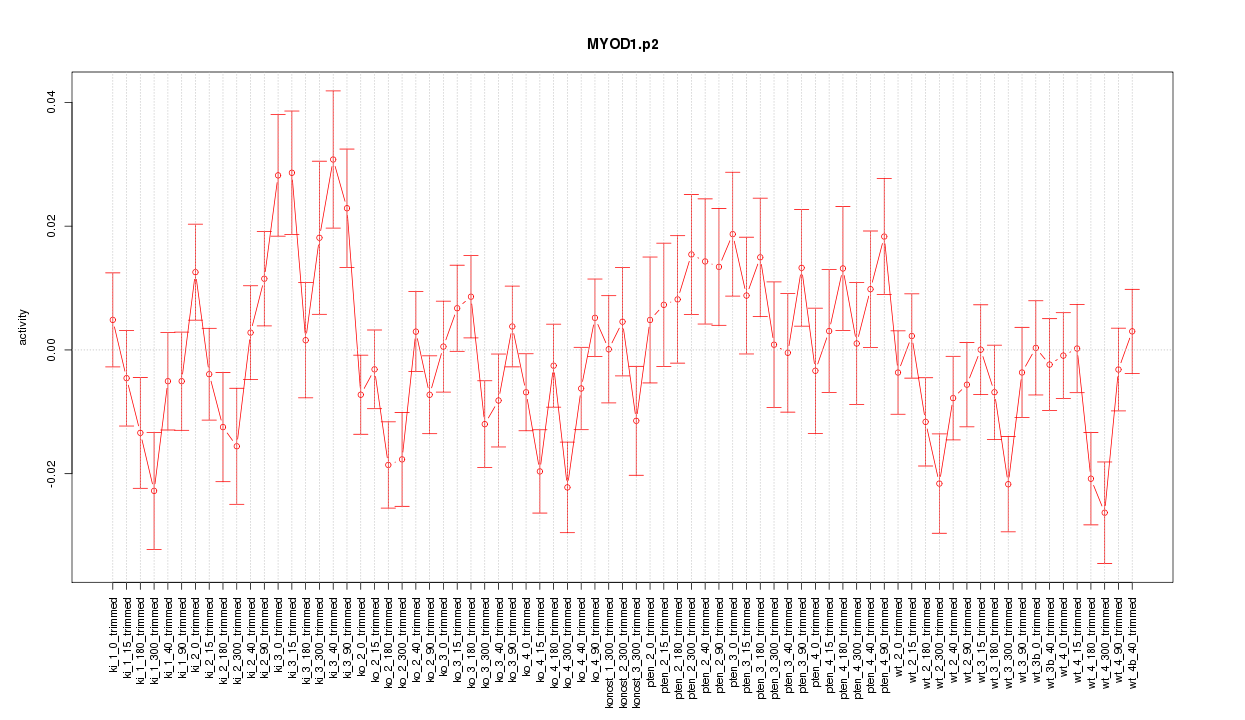
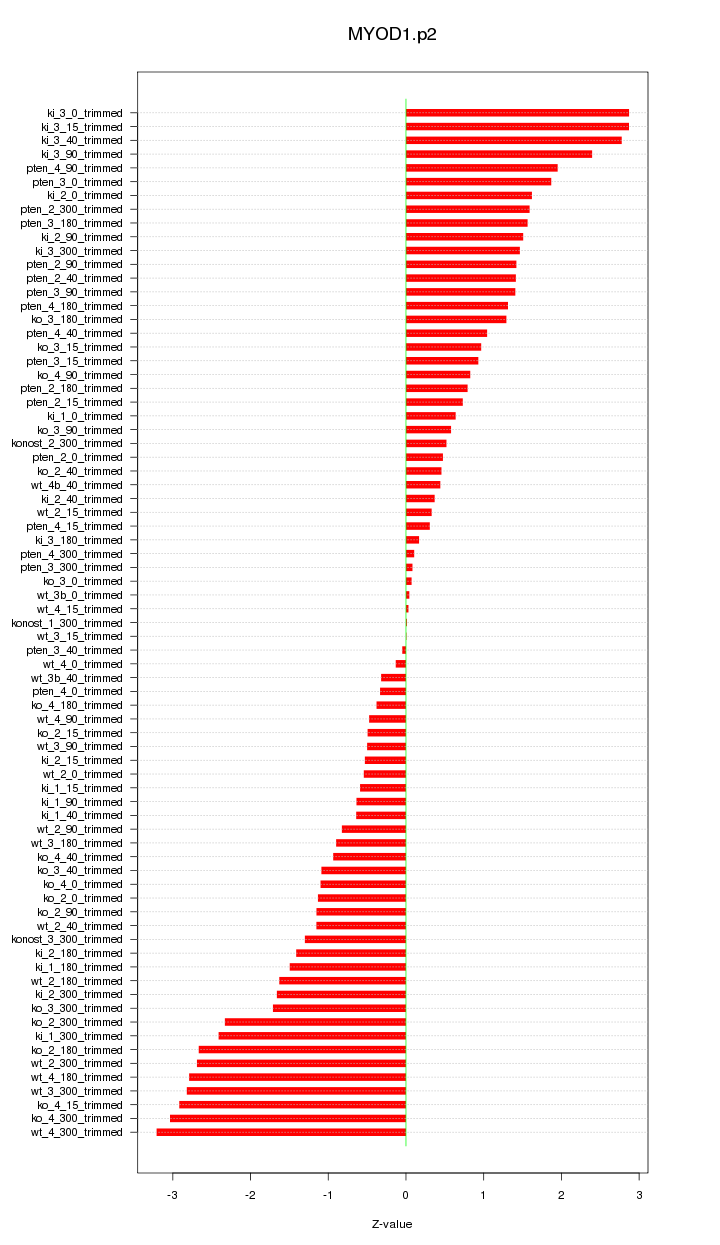
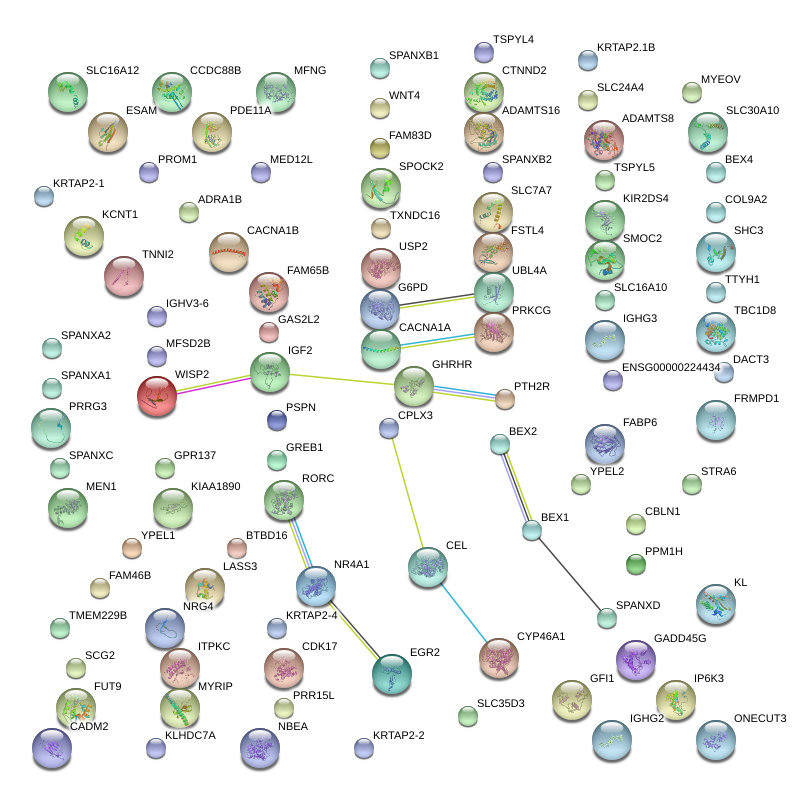
|
chr11_+_1860232
|
8.969
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chrX_+_140084755
|
6.403
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chrX_+_140096760
|
6.038
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chrX_-_130964633
|
5.964
|
|
LOC286467
|
family with sequence similarity 195, member A pseudogene
|
|
chr12_+_52416615
|
5.948
|
NM_001202233
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr5_+_159343739
|
5.855
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr11_+_69061599
|
5.662
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr1_-_27339326
|
5.409
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr9_+_37650996
|
5.404
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr15_-_74501309
|
5.123
|
NM_001142617
NM_001142619
NM_001142620
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr17_-_39203423
|
5.108
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr19_+_54926604
|
5.091
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr1_-_27339448
|
5.031
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr1_-_92951593
|
4.887
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr2_-_224467049
|
4.872
|
|
SCG2
|
secretogranin II
|
|
chr1_-_40782826
|
4.784
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr15_-_76352068
|
4.714
|
|
NRG4
|
neuregulin 4
|
|
chr1_-_40782924
|
4.694
|
NM_001852
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr17_-_39211462
|
4.640
|
NM_033032
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr17_-_46035109
|
4.441
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr13_+_35516390
|
4.441
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr2_-_224467095
|
4.426
|
|
SCG2
|
secretogranin II
|
|
chr2_+_209271555
|
4.361
|
NM_005048
|
PTH2R
|
parathyroid hormone 2 receptor
|
|
chr5_-_132948222
|
4.308
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr10_+_124030820
|
4.258
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chr2_-_224467142
|
4.171
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr1_-_40782981
|
4.135
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr10_-_64576123
|
4.083
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64576074
|
4.076
|
|
EGR2
|
early growth response 2
|
|
chr1_+_18807423
|
4.075
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr19_+_1753661
|
3.887
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr5_+_159614373
|
3.885
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr11_-_119234628
|
3.882
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_-_34079896
|
3.856
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr1_-_22469414
|
3.833
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr19_-_47164289
|
3.700
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chrX_-_140336628
|
3.632
|
NM_022661
|
SPANXC
|
SPANX family, member C
|
|
chr11_+_1860718
|
3.612
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr19_-_6375859
|
3.610
|
NM_004158
|
PSPN
|
persephin
|
|
chrX_-_102565805
|
3.587
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr19_+_41222915
|
3.583
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr1_-_92949500
|
3.520
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr1_-_92949355
|
3.518
|
NM_001127215
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr6_-_24911194
|
3.497
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr1_-_92949045
|
3.465
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr2_-_101767714
|
3.421
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr11_-_119252364
|
3.402
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr19_+_54385429
|
3.374
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr5_-_11904127
|
3.367
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr6_+_137243378
|
3.351
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chrX_+_150863729
|
3.349
|
NM_024082
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr14_-_67982020
|
3.342
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr19_+_41223282
|
3.331
|
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr9_+_92219926
|
3.315
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr14_-_107199126
|
3.250
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr6_-_33714681
|
3.221
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr11_+_64107687
|
3.216
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr22_-_22089975
|
3.210
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr3_+_85008031
|
3.206
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr6_+_111408780
|
3.204
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr3_+_39851028
|
3.195
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr12_-_63328663
|
3.122
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr20_+_43343523
|
3.116
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr11_-_2170792
|
3.115
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_-_23288912
|
3.112
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chrX_-_153775427
|
3.104
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr11_-_124632050
|
3.088
|
NM_138961
|
ESAM
|
endothelial cell adhesion molecule
|
|
chrX_+_102470003
|
3.083
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr9_+_140918042
|
3.075
|
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chrX_-_140672799
|
3.074
|
NM_013453
NM_145662
|
SPANXA1
SPANXA2
|
sperm protein associated with the nucleus, X-linked, family member A1
SPANX family, member A2
|
|
chr19_-_6199536
|
3.067
|
|
|
|
|
chr8_-_98290072
|
3.063
|
|
TSPYL5
|
TSPY-like 5
|
|
chr15_+_75118950
|
3.056
|
NM_001030005
|
CPLX3
|
complexin 3
|
|
chrX_-_140786561
|
3.032
|
NM_145665
NM_032417
|
SPANXE
SPANXD
SPANXA2
|
SPANX family, member E
SPANX family, member D
SPANX family, member A2
|
|
chr14_-_91526731
|
3.031
|
|
|
|
|
chr1_-_220101514
|
3.015
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr15_-_101084871
|
3.003
|
NM_178842
|
CERS3
|
ceramide synthase 3
|
|
chr22_-_37882214
|
2.988
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr2_+_24232952
|
2.974
|
NM_001080473
|
MFSD2B
|
major facilitator superfamily domain containing 2B
|
|
chr4_-_16085593
|
2.953
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr11_-_130298305
|
2.952
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr19_-_18391440
|
2.948
|
|
|
|
|
chr9_-_91793375
|
2.947
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr5_-_132948195
|
2.943
|
|
FSTL4
|
follistatin-like 4
|
|
chr20_+_37555055
|
2.929
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr1_-_151798552
|
2.927
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr11_-_119252267
|
2.920
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_11682836
|
2.911
|
NM_033090
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr16_-_49315682
|
2.906
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr5_+_5140442
|
2.901
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr9_+_135937364
|
2.899
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr13_+_33590477
|
2.886
|
NM_004795
|
KL
|
klotho
|
|
chr7_+_31003635
|
2.886
|
NM_000823
|
GHRHR
|
growth hormone releasing hormone receptor
|
|
chr3_+_150803448
|
2.884
|
|
MED12L
|
mediator complex subunit 12-like
|
|
chr2_-_178937472
|
2.881
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr6_+_96463844
|
2.862
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr14_+_92788924
|
2.851
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr10_-_91295310
|
2.846
|
NM_213606
|
SLC16A12
|
solute carrier family 16, member 12 (monocarboxylic acid transporter 12)
|
|
chr9_+_138594030
|
2.804
|
NM_020822
|
KCNT1
|
potassium channel, subfamily T, member 1
|
|
chr6_+_168841829
|
2.803
|
NM_001166412
NM_022138
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr1_-_32707198
|
2.803
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr14_+_100150596
|
2.786
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr10_-_73848766
|
2.771
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chrX_-_20009037
|
2.767
|
|
LOC729609
|
uncharacterized LOC729609
|
|
chr17_+_4981754
|
2.765
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr18_-_45935627
|
2.759
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr10_-_73848530
|
2.755
|
NM_001244950
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr19_-_47137938
|
2.721
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr1_+_55271735
|
2.716
|
NM_001110533
NM_152607
|
C1orf177
|
chromosome 1 open reading frame 177
|
|
chr2_+_79740059
|
2.701
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr15_+_47476225
|
2.698
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr7_+_140103842
|
2.685
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr10_+_99332197
|
2.677
|
NM_001129981
NM_020349
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle)
|
|
chr12_-_57351381
|
2.676
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr22_-_21386837
|
2.669
|
NM_004173
|
SLC7A4
|
solute carrier family 7 (orphan transporter), member 4
|
|
chr5_+_95067511
|
2.654
|
|
|
|
|
chr7_-_150675236
|
2.646
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr4_-_16085309
|
2.640
|
|
PROM1
|
prominin 1
|
|
chr11_+_2466171
|
2.628
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr8_-_143867931
|
2.627
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chrX_-_15402469
|
2.615
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chrX_+_150345055
|
2.599
|
NM_004224
|
GPR50
|
G protein-coupled receptor 50
|
|
chr3_-_197807541
|
2.596
|
|
ANKRD18DP
|
ankyrin repeat domain 18D, pseudogene
|
|
chr11_-_35441099
|
2.588
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_-_28303910
|
2.578
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chr16_+_56225250
|
2.567
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr1_-_161039413
|
2.566
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr1_-_16344438
|
2.552
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr17_+_4981721
|
2.549
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr7_+_100136784
|
2.549
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr6_+_168841873
|
2.519
|
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr10_+_112257624
|
2.517
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr16_+_23847271
|
2.500
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr9_-_139637354
|
2.490
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr11_-_130298502
|
2.475
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr19_-_460995
|
2.469
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr7_-_37488894
|
2.451
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_-_47734215
|
2.439
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr20_+_30407175
|
2.423
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr17_+_5972425
|
2.422
|
|
WSCD1
|
WSC domain containing 1
|
|
chr4_+_41362565
|
2.408
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_+_1203240
|
2.408
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr14_-_106237675
|
2.407
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr12_+_49524648
|
2.406
|
|
|
|
|
chr9_-_101470836
|
2.406
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr1_+_118148506
|
2.405
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr2_+_198669316
|
2.380
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr1_-_204120909
|
2.363
|
|
ETNK2
|
ethanolamine kinase 2
|
|
chr1_-_161039745
|
2.358
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr16_-_30124829
|
2.354
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr2_-_128433330
|
2.342
|
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr5_-_139422805
|
2.331
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chrX_-_152939256
|
2.321
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr22_+_21330361
|
2.318
|
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr19_+_15751706
|
2.315
|
NM_000896
NM_001199208
NM_001199209
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr11_+_7273400
|
2.305
|
|
SYT9
|
synaptotagmin IX
|
|
chr8_-_98290171
|
2.267
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr17_+_26800657
|
2.260
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr20_+_37554954
|
2.258
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr11_-_86383164
|
2.251
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr12_+_49372235
|
2.251
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr2_-_130956011
|
2.250
|
NM_207312
|
TUBA3E
|
tubulin, alpha 3e
|
|
chr2_+_234621637
|
2.246
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr19_+_56158953
|
2.245
|
NM_013301
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr4_-_159092509
|
2.245
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr11_-_2560814
|
2.243
|
|
|
|
|
chr2_+_234637772
|
2.241
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr2_-_109605668
|
2.240
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chrX_+_140677834
|
2.233
|
NM_013453
NM_145662
|
SPANXA1
SPANXA2
|
sperm protein associated with the nucleus, X-linked, family member A1
SPANX family, member A2
|
|
chr14_-_107083398
|
2.226
|
|
IGH@
IGHA1
IGHG1
|
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_-_153775064
|
2.198
|
NM_000402
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr8_-_124553448
|
2.185
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr7_+_100136845
|
2.167
|
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chrX_+_53111482
|
2.159
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr19_+_54926721
|
2.150
|
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr10_+_133918312
|
2.144
|
NM_001105521
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3
|
|
chr8_-_57123858
|
2.143
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr11_+_1874199
|
2.139
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chrX_-_153775002
|
2.132
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr16_+_2198687
|
2.122
|
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr8_-_57123827
|
2.121
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chrX_+_124453968
|
2.120
|
NM_001195272
|
LOC100129520
|
testis expressed sequence 13-like
|
|
chr2_+_239047362
|
2.118
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr16_+_330605
|
2.115
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr3_+_140396865
|
2.110
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr11_+_22214574
|
2.101
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr2_+_11680079
|
2.100
|
NM_148903
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr16_-_89007605
|
2.084
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr10_+_123872440
|
2.084
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr17_-_26692264
|
2.083
|
NM_001080837
|
SEBOX
|
SEBOX homeobox
|
|
chr11_+_117857123
|
2.078
|
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr10_-_73848278
|
2.074
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr17_-_39216343
|
2.071
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr1_-_9086403
|
2.066
|
NM_207420
|
SLC2A7
|
solute carrier family 2 (facilitated glucose transporter), member 7
|
|
chr1_+_201979689
|
2.065
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr4_-_2366530
|
2.059
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr12_-_63328857
|
2.057
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_-_39156106
|
2.056
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr1_-_9189087
|
2.055
|
NM_024980
|
GPR157
|
G protein-coupled receptor 157
|
|
chr16_+_84178864
|
2.053
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|