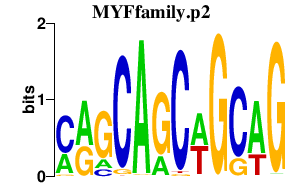
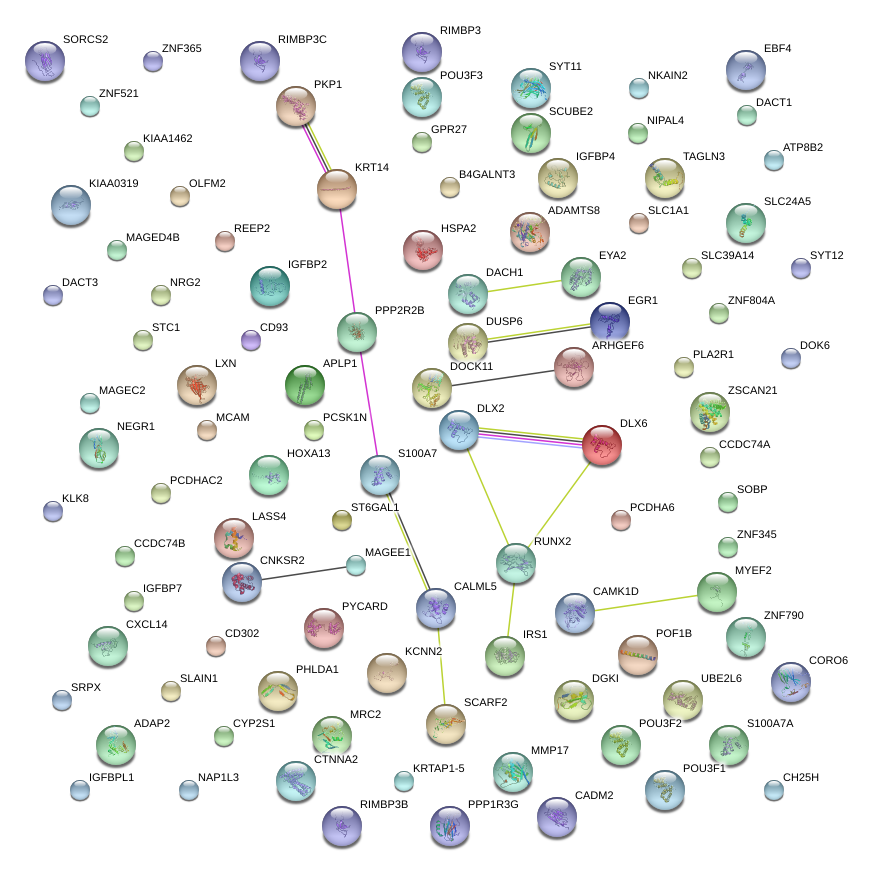
|
chr7_+_96635289
|
23.538
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr16_-_31214054
|
20.857
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chrX_-_38079980
|
19.506
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chrX_+_75648045
|
19.373
|
NM_020932
|
MAGEE1
|
melanoma antigen family E, 1
|
|
chr5_+_137801178
|
17.349
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr6_+_107811272
|
15.760
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_+_137774632
|
15.466
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chrX_-_84634664
|
13.501
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chrX_-_84634742
|
13.025
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr5_-_139422725
|
12.874
|
|
NRG2
|
neuregulin 2
|
|
chr19_+_41699076
|
12.861
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr6_+_45390284
|
12.662
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr15_-_48470453
|
12.167
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr6_+_45390376
|
12.011
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr2_+_217498082
|
11.943
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr5_+_137774897
|
11.907
|
|
REEP2
|
receptor accessory protein 2
|
|
chr2_+_217498205
|
11.574
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr2_-_160654668
|
11.461
|
|
CD302
|
CD302 molecule
|
|
chr4_-_57976516
|
11.435
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_57976550
|
11.392
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chrX_-_84634396
|
11.381
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr19_+_41699140
|
11.126
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr5_+_137801166
|
11.019
|
|
EGR1
|
early growth response 1
|
|
chr8_-_23712297
|
10.776
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chrX_-_48693934
|
10.572
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr16_-_31214003
|
10.223
|
|
PYCARD
|
PYD and CARD domain containing
|
|
chr22_+_21738039
|
9.631
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr6_+_99282597
|
9.610
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr3_+_71803200
|
9.591
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr22_-_20792107
|
9.585
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr10_+_64133915
|
9.581
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr13_-_72440143
|
9.217
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr5_+_140254930
|
9.162
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr15_-_48470319
|
9.053
|
|
MYEF2
|
myelin expression factor 2
|
|
chr2_-_227664330
|
9.046
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr20_+_45523247
|
8.886
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr5_-_146258143
|
8.844
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr19_+_36359346
|
8.516
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr17_+_60705051
|
8.386
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr10_-_90967048
|
8.332
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr12_-_89745753
|
8.277
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr5_+_140346289
|
8.267
|
NM_031883
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr12_-_76425375
|
8.200
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr2_+_132285469
|
8.064
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr19_-_51504799
|
7.925
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr17_+_60705042
|
7.895
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr20_+_45523505
|
7.840
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr13_+_78271820
|
7.782
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr10_-_30316338
|
7.731
|
|
KIAA1462
|
KIAA1462
|
|
chr17_-_27949814
|
7.647
|
|
CORO6
|
coronin 6
|
|
chr22_-_21905372
|
7.558
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr19_-_7701852
|
7.542
|
|
|
|
|
chr20_+_2673193
|
7.541
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr20_+_45523523
|
7.535
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr4_-_57976355
|
7.457
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr2_-_160654736
|
7.451
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr11_-_57335080
|
7.437
|
NM_004223
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr22_-_20792047
|
7.434
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr1_-_153433104
|
7.391
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr19_+_8274179
|
7.276
|
NM_024552
|
CERS4
|
ceramide synthase 4
|
|
chr19_-_36019194
|
7.268
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr12_+_132312917
|
7.113
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr15_+_48413168
|
7.072
|
NM_205850
|
SLC24A5
|
solute carrier family 24, member 5
|
|
chr12_+_569542
|
7.045
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr1_+_154301219
|
6.868
|
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr4_+_7194317
|
6.797
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chrX_+_117629871
|
6.712
|
NM_144658
|
DOCK11
|
dedicator of cytokinesis 11
|
|
chrX_-_135849356
|
6.711
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr9_-_38424422
|
6.697
|
NM_001007563
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1
|
|
chr5_+_113698015
|
6.650
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr11_+_66790815
|
6.494
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr12_-_89745997
|
6.396
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr7_-_27239724
|
6.382
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr12_-_76424474
|
6.376
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr14_+_65007418
|
6.277
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr13_-_72440557
|
6.241
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr9_+_4490193
|
6.231
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chrX_+_21392396
|
6.212
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr2_-_172967234
|
6.188
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr17_-_39743085
|
6.158
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr5_+_156887204
|
6.145
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr3_+_85008031
|
6.135
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr2_-_39187477
|
6.127
|
|
LOC375196
|
uncharacterized LOC375196
|
|
chr17_+_29248697
|
6.121
|
NM_018404
|
ADAP2
|
ArfGAP with dual PH domains 2
|
|
chr2_-_130902568
|
6.064
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr6_+_5085719
|
6.058
|
NM_001145115
|
PPP1R3G
|
protein phosphatase 1, regulatory subunit 3G
|
|
chr10_+_12391471
|
6.041
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr19_-_37341168
|
6.021
|
NM_001242801
|
ZNF790
|
zinc finger protein 790
|
|
chr19_-_37341688
|
5.959
|
NM_001242800
|
ZNF790
|
zinc finger protein 790
|
|
chr1_+_201252579
|
5.935
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr2_+_105471745
|
5.925
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr19_-_10047014
|
5.912
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr12_-_76425207
|
5.901
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chrX_-_92928566
|
5.851
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr5_+_113698304
|
5.846
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr5_+_140346053
|
5.833
|
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr6_+_124125068
|
5.793
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr14_+_65007163
|
5.756
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_+_185463235
|
5.717
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr17_+_38599651
|
5.713
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr1_-_72748147
|
5.705
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr10_+_12391728
|
5.703
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr18_-_22931970
|
5.666
|
|
ZNF521
|
zinc finger protein 521
|
|
chr5_-_134914447
|
5.657
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr17_-_39183453
|
5.461
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr10_-_5541401
|
5.398
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr20_-_23066828
|
5.383
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr19_+_37341259
|
5.366
|
NM_003419
|
ZNF345
|
zinc finger protein 345
|
|
chr3_+_111717585
|
5.347
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr3_-_158390266
|
5.324
|
NM_020169
|
LXN
|
latexin
|
|
chr11_-_9113092
|
5.309
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr2_-_160919120
|
5.300
|
NM_001007267
NM_001195641
NM_007366
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr3_+_186648273
|
5.260
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr2_+_79740059
|
5.224
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr17_+_60704747
|
5.167
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr11_-_119187811
|
5.106
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr7_-_137531585
|
5.098
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr21_+_46825291
|
5.094
|
|
|
|
|
chr14_+_59104736
|
5.087
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr11_-_130298502
|
5.078
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr3_+_111718006
|
5.046
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr8_+_31497267
|
5.045
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr4_-_6202251
|
4.954
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr2_+_37571752
|
4.945
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr1_+_171810611
|
4.914
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr5_-_139422805
|
4.887
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr19_+_51628136
|
4.852
|
NM_001198558
NM_014441
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9
|
|
chr6_+_31691120
|
4.795
|
NM_025260
NM_138272
NM_138273
NM_138274
NM_138275
NM_138277
|
C6orf25
|
chromosome 6 open reading frame 25
|
|
chr9_-_120177307
|
4.755
|
NM_014010
|
ASTN2
|
astrotactin 2
|
|
chr3_-_99833348
|
4.733
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_11918889
|
4.716
|
NM_002521
|
NPPB
|
natriuretic peptide B
|
|
chr17_+_19437140
|
4.712
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr5_+_52776479
|
4.712
|
|
FST
|
follistatin
|
|
chr3_+_3841079
|
4.701
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr6_-_32191779
|
4.699
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr3_-_64211111
|
4.661
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_-_160918935
|
4.641
|
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr19_+_37341724
|
4.637
|
NM_001242472
NM_001242474
|
ZNF345
|
zinc finger protein 345
|
|
chr8_-_140715234
|
4.594
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr5_+_156887026
|
4.586
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr2_+_176957448
|
4.573
|
NM_000523
|
HOXD13
|
homeobox D13
|
|
chr9_-_101470836
|
4.569
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr8_-_22550708
|
4.555
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr10_-_79397470
|
4.553
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr8_+_104383785
|
4.520
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr10_-_17496175
|
4.466
|
NM_001004470
|
ST8SIA6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6
|
|
chr11_-_627172
|
4.461
|
NM_021920
|
SCT
|
secretin
|
|
chr1_-_31712399
|
4.458
|
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chr2_-_47797469
|
4.401
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chrX_+_134555867
|
4.348
|
|
LINC00086
|
long intergenic non-protein coding RNA 86
|
|
chr19_-_39805975
|
4.344
|
NM_020862
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1
|
|
chr2_+_219724545
|
4.332
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr9_-_120177326
|
4.331
|
|
ASTN2
|
astrotactin 2
|
|
chr7_-_83278478
|
4.317
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr2_-_227663454
|
4.307
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr18_-_22932109
|
4.283
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr9_-_140063854
|
4.256
|
|
|
|
|
chr19_-_14048877
|
4.215
|
|
PODNL1
|
podocan-like 1
|
|
chr12_-_6665220
|
4.214
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chrX_-_153095996
|
4.210
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr11_+_94277566
|
4.207
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr5_-_147162264
|
4.203
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr11_-_130298305
|
4.198
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr12_+_103352021
|
4.192
|
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr10_-_79397546
|
4.175
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr10_-_79397394
|
4.168
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr18_+_10454542
|
4.157
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr1_+_221053000
|
4.135
|
|
HLX
|
H2.0-like homeobox
|
|
chr3_+_139654206
|
4.135
|
|
CLSTN2
|
calsyntenin 2
|
|
chr3_+_139654001
|
4.132
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr19_-_47137938
|
4.126
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr12_-_95044205
|
4.117
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr2_-_193059599
|
4.102
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr16_-_4588738
|
4.090
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr8_+_1950304
|
4.054
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr14_-_27066959
|
4.051
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr17_+_7452374
|
4.047
|
NM_172089
NM_003809
|
TNFSF12-TNFSF13
TNFSF12
|
TNFSF12-TNFSF13 readthrough
tumor necrosis factor (ligand) superfamily, member 12
|
|
chr2_-_172967627
|
4.030
|
|
DLX2
|
distal-less homeobox 2
|
|
chr2_+_30454396
|
4.026
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr9_+_33795558
|
4.023
|
NM_002771
|
PRSS3
|
protease, serine, 3
|
|
chr21_-_28217272
|
4.006
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr17_+_61554420
|
4.001
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr16_-_14788429
|
3.980
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr19_+_45409003
|
3.976
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr11_-_12030128
|
3.947
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_-_31712733
|
3.946
|
NM_024522
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chr5_+_52776263
|
3.931
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr22_+_24115035
|
3.909
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr3_+_186648515
|
3.903
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr2_-_48982809
|
3.862
|
NM_000233
|
LHCGR
|
luteinizing hormone/choriogonadotropin receptor
|
|
chr2_-_172967014
|
3.843
|
|
DLX2
|
distal-less homeobox 2
|
|
chr14_-_75078669
|
3.815
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr5_-_141993691
|
3.807
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_115632047
|
3.793
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chrX_-_107979606
|
3.777
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr10_+_52834233
|
3.761
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr5_-_147162175
|
3.758
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_+_140770412
|
3.729
|
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr19_-_15343194
|
3.723
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr15_+_81426605
|
3.723
|
NM_173528
|
C15orf26
|
chromosome 15 open reading frame 26
|