|
chr3_+_71803200
|
20.687
|
NM_018971
|
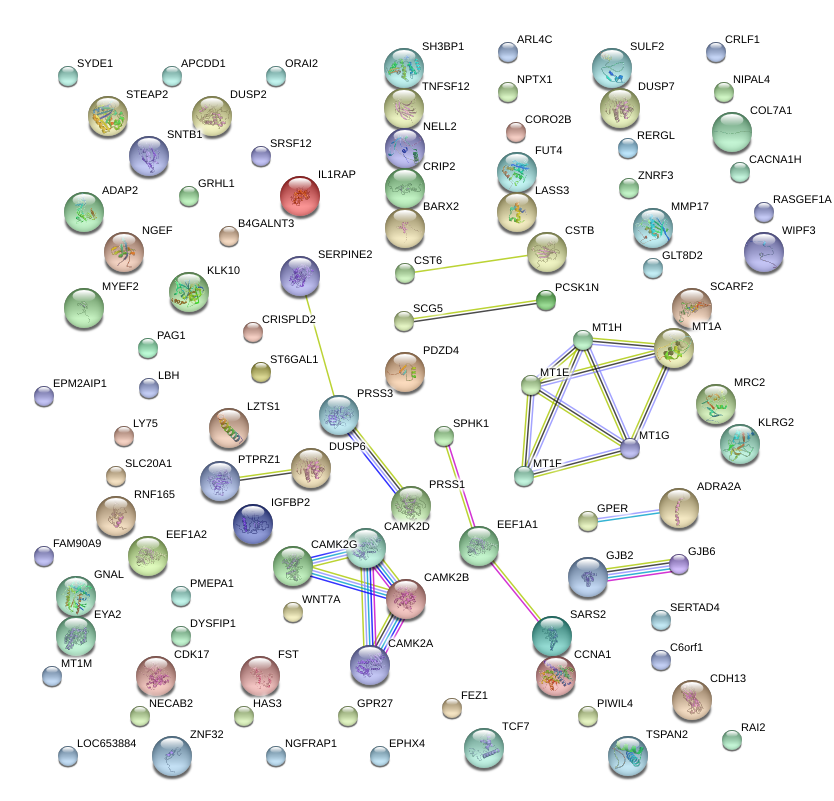
GPR27
|
G protein-coupled receptor 27
|
|
chr13_-_20767100
|
17.582
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr19_+_15218141
|
16.401
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr13_-_20806533
|
15.058
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_+_92495484
|
14.734
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr2_+_217498082
|
14.171
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr10_-_43762366
|
12.612
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chrX_+_102631408
|
12.404
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr13_-_20767036
|
12.089
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr7_-_44365019
|
11.512
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr17_+_60705051
|
11.433
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr17_+_60705042
|
11.134
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr2_+_217498205
|
10.515
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr11_+_129245834
|
10.429
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr13_+_37006408
|
10.336
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr2_-_224903933
|
10.033
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr13_+_37005966
|
9.288
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr10_-_44144151
|
9.267
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr15_-_48470453
|
9.117
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr17_+_74380676
|
9.051
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr8_-_121824287
|
9.046
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr12_+_569542
|
8.887
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr12_-_45270157
|
8.820
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_48632592
|
8.723
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr7_+_121513158
|
8.586
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr3_+_190231839
|
8.498
|
NM_001167928
NM_001167929
NM_001167930
NM_001167931
NM_002182
NM_134470
|
IL1RAP
|
interleukin 1 receptor accessory protein
|
|
chr12_+_132312917
|
8.230
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr5_+_133450353
|
8.198
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr8_-_82024277
|
8.049
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr18_+_11689135
|
7.915
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr12_-_89745997
|
7.755
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chrX_+_102632108
|
7.535
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chrX_-_48693934
|
7.534
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr7_+_29846112
|
7.527
|
NM_001080529
|
WIPF3
|
WAS/WASL interacting protein family, member 3
|
|
chr3_+_186648515
|
7.426
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr16_+_56703725
|
7.264
|
NM_005951
|
MT1H
|
metallothionein 1H
|
|
chr20_+_45523247
|
7.246
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr7_+_102074002
|
7.219
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr8_-_121824268
|
7.050
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr12_-_45270071
|
7.037
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr5_+_52776263
|
6.912
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr17_+_7452374
|
6.883
|
NM_172089
NM_003809
|
TNFSF12-TNFSF13
TNFSF12
|
TNFSF12-TNFSF13 readthrough
tumor necrosis factor (ligand) superfamily, member 12
|
|
chr19_-_18717467
|
6.812
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr16_+_84853585
|
6.778
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr17_-_78450247
|
6.771
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr20_+_45523523
|
6.684
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr3_-_13921617
|
6.639
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr20_+_45523505
|
6.623
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr2_-_160761192
|
6.496
|
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr19_-_51522953
|
6.418
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr19_-_18717631
|
6.417
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr16_-_56701904
|
6.393
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr22_-_20792047
|
6.392
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr3_-_52090090
|
6.357
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr7_+_121513210
|
6.340
|
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr2_-_233792825
|
6.276
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr17_+_60704747
|
6.150
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr2_+_113403433
|
6.097
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr20_-_56284945
|
6.095
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr6_-_89827605
|
6.054
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr20_-_46414636
|
5.990
|
NM_018837
|
SULF2
|
sulfatase 2
|
|
chr14_+_105941061
|
5.985
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr7_+_89840999
|
5.968
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr17_+_29248697
|
5.964
|
NM_018404
|
ADAP2
|
ArfGAP with dual PH domains 2
|
|
chr18_+_10454620
|
5.901
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr10_-_44144266
|
5.844
|
NM_006973
|
ZNF32
|
zinc finger protein 32
|
|
chrX_-_153095996
|
5.834
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr5_+_156887204
|
5.824
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr11_+_65779461
|
5.743
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr2_-_235405692
|
5.727
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_115632047
|
5.724
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr1_+_210406194
|
5.717
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr11_-_125366048
|
5.668
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr3_-_45267804
|
5.646
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr12_-_45270632
|
5.617
|
NM_001145107
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr15_-_101084871
|
5.615
|
NM_178842
|
CERS3
|
ceramide synthase 3
|
|
chr7_+_1126442
|
5.585
|
NM_001039966
NM_001505
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr17_+_74380943
|
5.577
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr22_-_20792107
|
5.548
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr9_+_33750463
|
5.546
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chrX_+_102631267
|
5.500
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr8_+_121823538
|
5.460
|
|
|
|
|
chr16_+_56666533
|
5.407
|
NM_176870
|
MT1M
|
metallothionein 1M
|
|
chr15_+_68871285
|
5.403
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr16_+_82660573
|
5.375
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr15_+_32933869
|
5.350
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr12_-_45269953
|
5.347
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr5_-_138730884
|
5.331
|
NM_001161546
|
C5orf65
|
chromosome 5 open reading frame 65
|
|
chr12_-_93964636
|
5.308
|
|
|
|
|
chr16_+_69139466
|
5.267
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr16_+_1203240
|
5.265
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr20_-_62130390
|
5.221
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr2_-_96810775
|
5.157
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr8_-_121824015
|
5.139
|
|
|
|
|
chrX_-_17879355
|
5.033
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr16_+_84002099
|
5.021
|
NM_019065
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr2_+_10091791
|
5.019
|
NM_198182
|
GRHL1
|
grainyhead-like 1 (Drosophila)
|
|
chr18_+_43913906
|
5.014
|
|
RNF165
|
ring finger protein 165
|
|
chr7_-_139168408
|
5.012
|
NM_198508
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2
|
|
chr2_+_30454396
|
4.979
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr22_+_38035683
|
4.969
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr11_+_94277005
|
4.935
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr22_+_29279573
|
4.921
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr22_+_38035718
|
4.920
|
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr6_-_110679474
|
4.908
|
NM_001123364
|
C6orf186
|
chromosome 6 open reading frame 186
|
|
chr19_-_51523272
|
4.876
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr15_+_73976537
|
4.827
|
|
|
|
|
chr15_+_25200069
|
4.808
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr10_-_92617417
|
4.803
|
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr11_+_1411089
|
4.781
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr15_+_69591272
|
4.773
|
NM_017705
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chrX_+_68725077
|
4.761
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr16_+_82660641
|
4.733
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr20_-_4804067
|
4.719
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr1_-_6321034
|
4.677
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr11_-_75062650
|
4.670
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr11_+_35639734
|
4.640
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr19_+_21579884
|
4.608
|
NM_001076678
NM_145326
NM_175910
|
ZNF493
|
zinc finger protein 493
|
|
chr3_-_16555128
|
4.604
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr2_+_241375207
|
4.590
|
|
GPC1
|
glypican 1
|
|
chr8_-_145515014
|
4.586
|
|
BOP1
|
block of proliferation 1
|
|
chr17_-_46655544
|
4.582
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr2_+_132285469
|
4.567
|
NM_138770
|
CCDC74A
|
coiled-coil domain containing 74A
|
|
chr22_+_38201232
|
4.557
|
|
H1F0
|
H1 histone family, member 0
|
|
chr10_-_92617670
|
4.556
|
NM_000872
NM_019859
NM_019860
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr9_+_17578952
|
4.545
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr8_+_1712011
|
4.536
|
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr1_+_15250578
|
4.525
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr7_+_73082320
|
4.516
|
|
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae)
|
|
chr2_-_96811138
|
4.510
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr3_-_53880403
|
4.482
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr3_-_183543295
|
4.477
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr4_+_42399855
|
4.458
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr3_-_38691118
|
4.451
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr18_-_5543967
|
4.442
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr9_+_140772228
|
4.426
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr15_-_26108236
|
4.414
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr7_+_29234325
|
4.404
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr1_-_85358851
|
4.392
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr12_-_133464197
|
4.381
|
NM_001161344
NM_001161345
NM_001161346
NM_001161347
NM_018223
|
CHFR
|
checkpoint with forkhead and ring finger domains
|
|
chrX_+_152760346
|
4.381
|
NM_001711
|
BGN
|
biglycan
|
|
chr14_-_99737554
|
4.363
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr2_-_110873353
|
4.362
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr22_+_38201113
|
4.335
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr6_+_160148485
|
4.333
|
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr16_+_27325244
|
4.319
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr8_+_146277789
|
4.310
|
NM_023080
|
C8orf33
|
chromosome 8 open reading frame 33
|
|
chr12_-_76425375
|
4.308
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr5_+_176873795
|
4.289
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chrX_-_43741593
|
4.285
|
NM_000898
|
MAOB
|
monoamine oxidase B
|
|
chr1_+_226250413
|
4.278
|
|
H3F3A
|
H3 histone, family 3A
|
|
chr16_+_1203737
|
4.267
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr13_-_77460432
|
4.254
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr12_-_89746244
|
4.247
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr16_+_27325269
|
4.239
|
|
IL4R
|
interleukin 4 receptor
|
|
chr15_+_80696569
|
4.233
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr21_-_43299407
|
4.232
|
NM_001040424
NM_022115
|
PRDM15
|
PR domain containing 15
|
|
chr3_-_185542744
|
4.228
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr12_-_89745753
|
4.226
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_-_100721128
|
4.215
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr17_-_27949814
|
4.188
|
|
CORO6
|
coronin 6
|
|
chr9_-_38069082
|
4.180
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr18_+_77155966
|
4.170
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr13_-_80915041
|
4.153
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr11_+_125774271
|
4.149
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr20_-_10654139
|
4.128
|
|
JAG1
|
jagged 1
|
|
chr16_+_56716378
|
4.123
|
NM_005952
|
MT1X
|
metallothionein 1X
|
|
chr13_-_77460369
|
4.118
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr19_-_18717577
|
4.117
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr20_-_55841671
|
4.112
|
NM_001719
|
BMP7
|
bone morphogenetic protein 7
|
|
chr14_+_65007418
|
4.100
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr16_+_69141442
|
4.078
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr9_-_113800243
|
4.068
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chrX_-_8700170
|
4.053
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr16_-_89007605
|
4.052
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr19_-_12251092
|
4.042
|
NM_001203250
NM_021143
|
ZNF20
|
zinc finger protein 20
|
|
chr7_+_102073953
|
4.037
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chrX_-_64254592
|
4.025
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr18_+_77155927
|
4.003
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_-_45267620
|
3.967
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr6_-_170600165
|
3.962
|
|
|
|
|
chr1_+_9648931
|
3.959
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr10_+_12391728
|
3.937
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr1_-_200992824
|
3.936
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr11_-_12030128
|
3.927
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr7_-_29186072
|
3.925
|
NM_019029
NM_031311
|
CPVL
|
carboxypeptidase, vitellogenic-like
|
|
chr1_+_154298026
|
3.919
|
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr12_-_76425207
|
3.915
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr12_-_89746238
|
3.878
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr20_-_32274070
|
3.835
|
|
E2F1
|
E2F transcription factor 1
|
|
chr1_+_154298034
|
3.818
|
NM_001005855
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr9_-_113800980
|
3.805
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr12_-_120806893
|
3.796
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr19_+_589849
|
3.796
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr1_-_62785067
|
3.782
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr19_-_51512803
|
3.755
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr2_-_204399906
|
3.747
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr11_-_67141008
|
3.735
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr3_+_36421944
|
3.730
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr17_-_80656461
|
3.715
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|