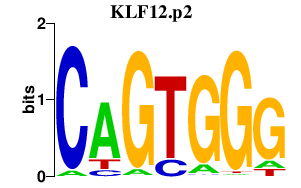
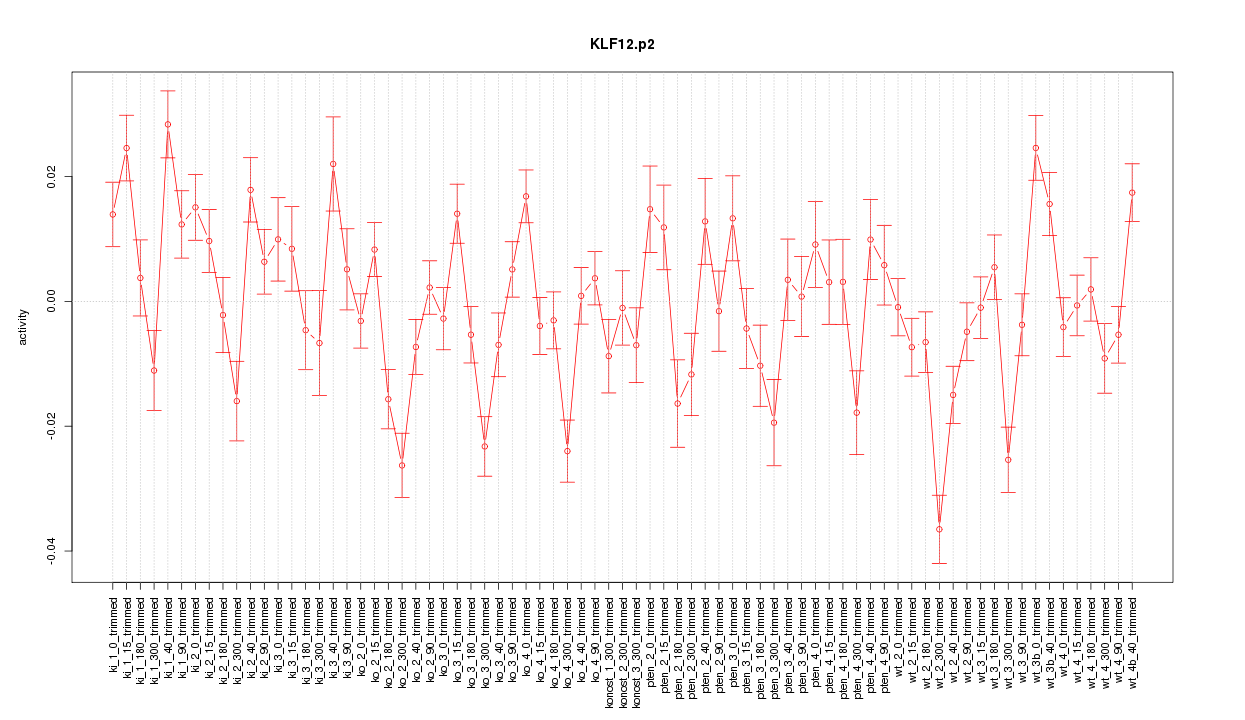
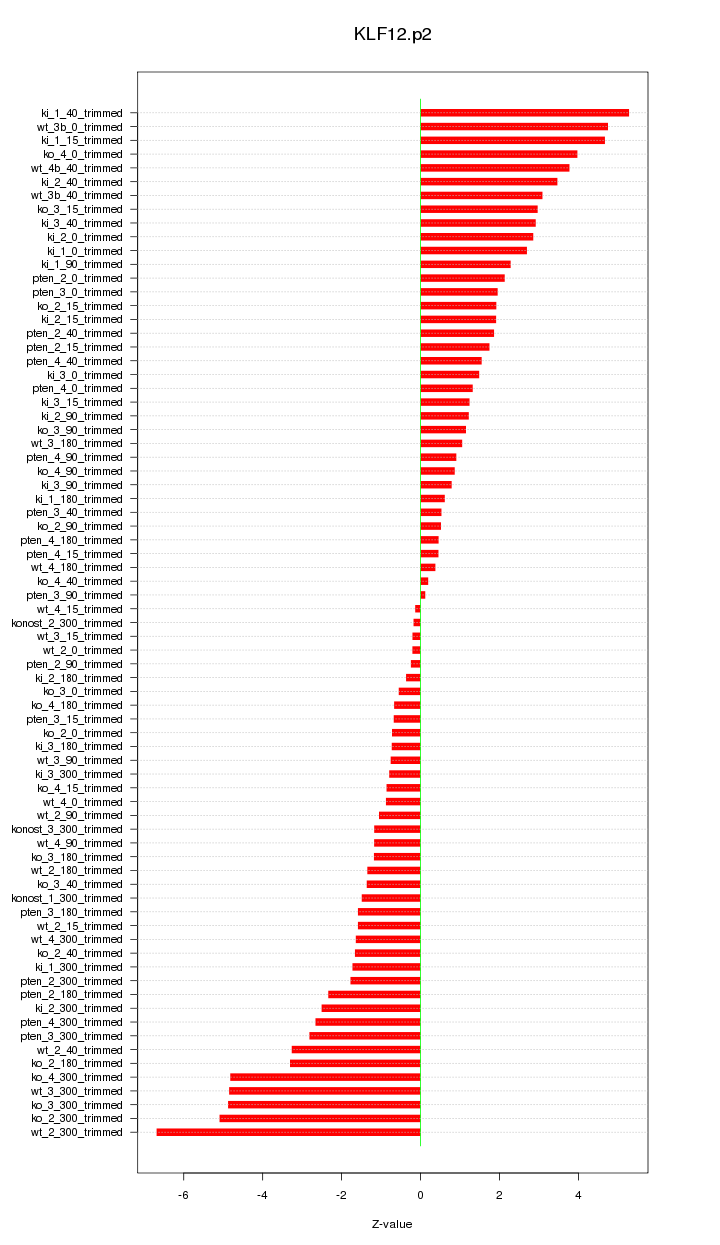
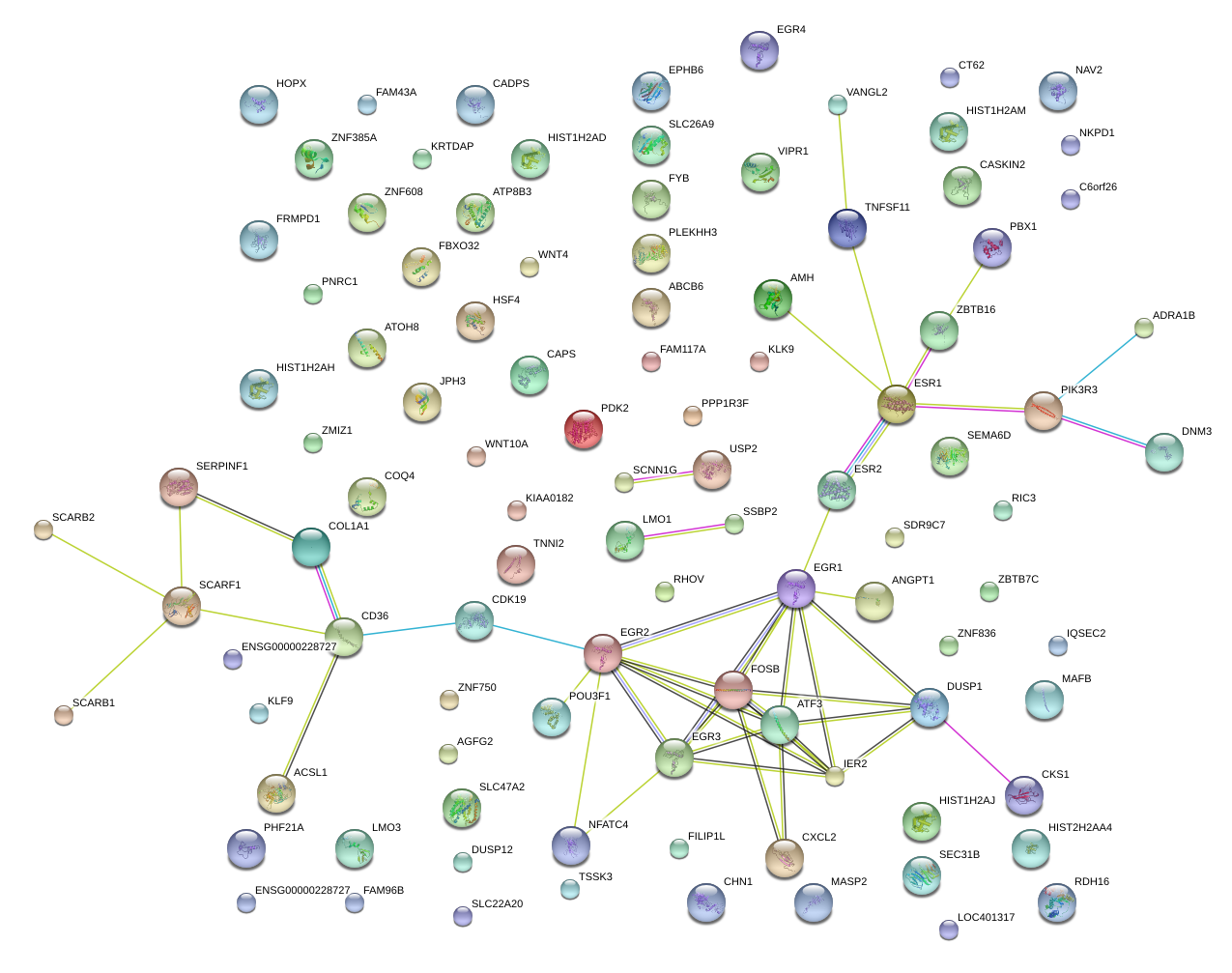
|
chr5_+_137801166
|
20.666
|
|
EGR1
|
early growth response 1
|
|
chr5_+_137801178
|
19.806
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr19_-_35981345
|
16.222
|
NM_001244847
NM_207392
|
KRTDAP
|
keratinocyte differentiation-associated protein
|
|
chr2_-_73520448
|
13.432
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr17_-_80797889
|
9.496
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr8_-_22550057
|
9.048
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr3_+_42544131
|
8.860
|
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr5_+_159343739
|
8.793
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr1_+_164528914
|
8.637
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr17_+_1665342
|
8.619
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr18_-_45663667
|
8.241
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr10_-_64576074
|
7.864
|
|
EGR2
|
early growth response 2
|
|
chr13_+_43136871
|
7.739
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr17_+_1665306
|
7.176
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr7_+_100136784
|
7.166
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr8_-_22550456
|
7.163
|
|
EGR3
|
early growth response 3
|
|
chr17_+_1665281
|
7.141
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr7_+_100136845
|
7.124
|
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr1_-_46598250
|
7.079
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr1_-_32827843
|
7.007
|
NM_001167676
|
LOC100128071
|
uncharacterized LOC100128071
|
|
chr2_+_219745243
|
6.922
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr19_+_2249112
|
6.798
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr1_+_32827797
|
6.469
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr17_-_48265912
|
6.298
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_22469414
|
6.234
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr19_+_45971249
|
6.146
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr7_+_142552547
|
6.045
|
NM_004445
|
EPHB6
|
EPH receptor B6
|
|
chr2_+_85980600
|
5.931
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr4_-_57522469
|
5.837
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr3_+_194407412
|
5.806
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr12_-_57351381
|
5.720
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr14_+_95623991
|
5.662
|
|
DICER1-AS
|
DICER1 antisense RNA (non-protein coding)
|
|
chr11_+_113930430
|
5.609
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr5_+_133842276
|
5.574
|
|
|
|
|
chr17_+_1665233
|
5.522
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr11_+_64981310
|
5.455
|
NM_001004326
|
SLC22A20
|
solute carrier family 22, member 20
|
|
chr3_-_169482847
|
5.447
|
|
TERC
|
telomerase RNA component
|
|
chr19_-_51512803
|
5.364
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr2_-_220083394
|
5.335
|
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr9_-_73028278
|
5.316
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr6_-_111136616
|
5.311
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_+_13263890
|
5.260
|
|
IER2
|
immediate early response 2
|
|
chr13_+_43148182
|
5.251
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chrX_+_49126326
|
5.248
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr10_+_81002836
|
5.192
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr11_+_113930296
|
5.134
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_22549901
|
5.081
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr11_-_8190535
|
5.050
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr3_+_42544094
|
5.018
|
NM_001251883
NM_001251884
NM_001251885
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr15_-_71407800
|
4.970
|
NM_001102658
|
CT62
|
cancer/testis antigen 62
|
|
chr11_-_8285299
|
4.941
|
NM_002315
|
LMO1
|
LIM domain only 1 (rhombotin 1)
|
|
chr20_-_39317875
|
4.940
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr11_-_2018334
|
4.928
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr2_-_220083658
|
4.908
|
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr1_+_160370363
|
4.907
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr6_+_31730772
|
4.856
|
NM_001039651
|
C6orf26
|
chromosome 6 open reading frame 26
|
|
chr16_+_67198678
|
4.854
|
|
HSF4
|
heat shock transcription factor 4
|
|
chr10_-_64576123
|
4.828
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chrX_+_49126299
|
4.821
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr7_+_28448970
|
4.800
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr2_-_220083699
|
4.764
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr5_-_81046858
|
4.758
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr5_-_124080773
|
4.751
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr11_-_2018446
|
4.698
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr17_-_1548992
|
4.637
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr17_-_19619878
|
4.607
|
NM_001099646
NM_152908
|
SLC47A2
|
solute carrier family 47, member 2
|
|
chr5_-_172198160
|
4.600
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr4_-_57522059
|
4.597
|
|
HOPX
|
HOP homeobox
|
|
chr11_-_119252364
|
4.539
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_-_48266572
|
4.529
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr4_-_185726865
|
4.502
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr5_-_39203061
|
4.468
|
|
FYB
|
FYN binding protein
|
|
chr4_-_74964836
|
4.444
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr6_-_111136506
|
4.441
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_-_205912546
|
4.431
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr11_-_2018568
|
4.403
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr6_-_27782517
|
4.360
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr10_-_102279590
|
4.359
|
NM_015490
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chr11_-_46142955
|
4.347
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chrX_-_53350521
|
4.325
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr17_-_40828441
|
4.295
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr5_-_81046943
|
4.285
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr19_-_45663407
|
4.263
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr15_+_47476225
|
4.254
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr15_-_41166336
|
4.238
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr2_-_175712276
|
4.213
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr11_+_1860232
|
4.170
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr11_-_2017822
|
4.160
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr17_+_48172508
|
4.147
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr19_-_39402797
|
4.140
|
NM_001243212
|
LOC643669
|
uncharacterized LOC643669
|
|
chr16_+_67198583
|
4.120
|
|
HSF4
|
heat shock transcription factor 4
|
|
chr12_-_54778298
|
4.111
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr19_+_5914192
|
4.110
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr14_+_24837225
|
4.074
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr11_+_20070609
|
4.056
|
|
NAV2
|
neuron navigator 2
|
|
chr9_+_37650996
|
4.047
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr1_+_164528586
|
4.021
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr16_+_85646923
|
4.005
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr6_-_111136407
|
4.001
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr4_-_74964996
|
3.999
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr1_-_11107150
|
3.994
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr6_+_152126807
|
3.961
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr1_+_171810611
|
3.958
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr6_+_89790464
|
3.957
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr16_+_67198637
|
3.933
|
|
HSF4
|
heat shock transcription factor 4
|
|
chr1_-_38512446
|
3.852
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr3_-_99833348
|
3.840
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_-_57328044
|
3.836
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr1_+_212781969
|
3.835
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr8_-_124553253
|
3.828
|
|
FBXO32
|
F-box protein 32
|
|
chr9_+_131085052
|
3.818
|
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chrX_+_49126480
|
3.802
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr8_-_108510094
|
3.801
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr16_+_23194037
|
3.779
|
NM_001039
|
SCNN1G
|
sodium channel, nonvoltage-gated 1, gamma
|
|
chr17_-_47841517
|
3.778
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr7_+_80275967
|
3.764
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr16_+_87636492
|
3.742
|
NM_020655
|
JPH3
|
junctophilin 3
|
|
chr19_-_1812198
|
3.728
|
NM_001178002
NM_138813
|
ATP8B3
|
ATPase, aminophospholipid transporter, class I, type 8B, member 3
|
|
chr6_+_89790412
|
3.721
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chrX_-_15288172
|
3.713
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr12_-_54778456
|
3.700
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr9_+_135937364
|
3.698
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr3_-_49459870
|
3.676
|
|
AMT
|
aminomethyltransferase
|
|
chr17_-_34079896
|
3.660
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr11_+_7535213
|
3.647
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr10_+_23983674
|
3.641
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr11_-_119252267
|
3.637
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_-_8055684
|
3.622
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr9_+_139871954
|
3.598
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr1_+_209941894
|
3.590
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr2_+_11680079
|
3.570
|
NM_148903
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr22_-_30685485
|
3.562
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr11_-_2018703
|
3.509
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr9_-_130533509
|
3.506
|
NM_001252334
NM_005489
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr1_-_85741992
|
3.502
|
|
BCL10
|
B-cell CLL/lymphoma 10
|
|
chr1_+_48688356
|
3.498
|
NM_001011547
NM_001135181
|
SLC5A9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9
|
|
chr8_-_145060594
|
3.488
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr6_-_11382501
|
3.443
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr17_-_47841468
|
3.418
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr1_+_109792640
|
3.416
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr20_-_32307889
|
3.409
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr17_-_38978824
|
3.408
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr4_-_149363498
|
3.407
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr13_-_67804432
|
3.406
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr8_-_145060563
|
3.405
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr5_-_95768660
|
3.398
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr19_+_50380681
|
3.396
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr19_+_49838700
|
3.392
|
|
CD37
|
CD37 molecule
|
|
chr19_+_2476122
|
3.392
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr11_+_113931228
|
3.381
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr17_+_42634598
|
3.374
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr18_+_44497512
|
3.370
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr17_+_48262816
|
3.362
|
|
|
|
|
chr12_-_53074172
|
3.361
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr9_+_131084745
|
3.351
|
NM_016035
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chr11_-_128807712
|
3.331
|
NM_001195194
|
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1
|
|
chr14_-_67982020
|
3.330
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr5_-_176936857
|
3.330
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr17_-_74533623
|
3.330
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr17_+_26699053
|
3.321
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr8_-_20112691
|
3.318
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr1_+_212738675
|
3.314
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_123357503
|
3.295
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr20_-_2821245
|
3.278
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr22_-_40859423
|
3.268
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr11_+_64120348
|
3.261
|
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr17_-_48262900
|
3.246
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr8_-_48650683
|
3.245
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr20_+_306205
|
3.241
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr3_-_87040268
|
3.230
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr12_-_27235454
|
3.211
|
NM_001080406
|
C12orf71
|
chromosome 12 open reading frame 71
|
|
chr22_-_39268214
|
3.194
|
|
CBX6
|
chromobox homolog 6
|
|
chr17_+_73629513
|
3.187
|
NM_001162995
|
C17orf109
|
chromosome 17 open reading frame 109
|
|
chr10_+_5566923
|
3.181
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr22_-_39268230
|
3.177
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr2_+_241375207
|
3.164
|
|
GPC1
|
glypican 1
|
|
chr12_-_54694717
|
3.145
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr11_-_67442086
|
3.141
|
NM_000695
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr19_-_46272102
|
3.139
|
|
|
|
|
chr9_+_116263706
|
3.135
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr17_+_26699093
|
3.133
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr1_-_221915393
|
3.123
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr6_-_111136330
|
3.123
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr15_-_40401041
|
3.123
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr10_+_23728197
|
3.092
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr16_+_69984809
|
3.091
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr2_+_11682836
|
3.085
|
NM_033090
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr11_-_85469043
|
3.083
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr14_-_91526912
|
3.072
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr2_+_166428845
|
3.053
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr20_-_2821120
|
3.052
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr6_+_99872769
|
3.022
|
|
|
|
|
chr5_+_131705353
|
2.989
|
NM_003060
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5
|
|
chr13_+_97999289
|
2.977
|
|
|
|
|
chr15_+_66994669
|
2.975
|
NM_005585
|
SMAD6
|
SMAD family member 6
|
|
chr1_-_149812733
|
2.972
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr4_-_74904329
|
2.963
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr19_-_49140569
|
2.963
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr2_+_203500211
|
2.958
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chrX_-_48828250
|
2.949
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|