|
chr19_+_45971249
|
28.331
|
NM_001114171
NM_006732
|
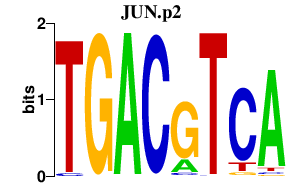
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr10_-_64576123
|
15.940
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64576074
|
15.627
|
|
EGR2
|
early growth response 2
|
|
chr1_+_212782103
|
12.053
|
|
ATF3
|
activating transcription factor 3
|
|
chr8_-_22550708
|
9.854
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr2_-_73520448
|
9.601
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr2_-_224467049
|
9.223
|
|
SCG2
|
secretogranin II
|
|
chr5_-_172198160
|
8.618
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr16_+_56225250
|
8.555
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr16_-_79634610
|
8.143
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr19_+_1753661
|
7.953
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr7_+_28448970
|
7.878
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr2_-_224467095
|
7.690
|
|
SCG2
|
secretogranin II
|
|
chr5_-_132948222
|
7.546
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr1_+_212738675
|
7.342
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr16_+_56225301
|
7.311
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr11_-_119234859
|
7.298
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_-_1593149
|
7.126
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr9_+_102584136
|
7.003
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr21_-_44846927
|
6.912
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr2_-_224467142
|
6.670
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr1_-_59249687
|
6.531
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr19_-_39402797
|
6.486
|
NM_001243212
|
LOC643669
|
uncharacterized LOC643669
|
|
chr11_-_119234628
|
5.900
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_162346579
|
5.855
|
NM_182581
|
C1orf111
|
chromosome 1 open reading frame 111
|
|
chr22_-_44258210
|
5.831
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr19_+_46733008
|
5.827
|
NM_198541
|
IGFL1
|
IGF-like family member 1
|
|
chr1_-_16482426
|
5.669
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr19_+_49375638
|
5.355
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr19_+_49375677
|
5.340
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr14_-_77494974
|
5.281
|
NM_024496
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr19_+_49375679
|
5.267
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr12_-_123380682
|
5.250
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr1_+_212781969
|
5.102
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr4_-_23891608
|
5.005
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr10_-_1779669
|
4.802
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr1_+_152956563
|
4.742
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr10_+_5566923
|
4.734
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr17_-_8055684
|
4.677
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr19_+_13263890
|
4.618
|
|
IER2
|
immediate early response 2
|
|
chr5_+_137801166
|
4.604
|
|
EGR1
|
early growth response 1
|
|
chr20_+_43344005
|
4.499
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr22_+_38597938
|
4.493
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr12_-_123380628
|
4.427
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr2_+_241375207
|
4.419
|
|
GPC1
|
glypican 1
|
|
chr10_+_123923104
|
4.307
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_+_228678556
|
4.207
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr9_+_130860769
|
4.146
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr22_+_38598065
|
4.121
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr10_+_123922940
|
4.038
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr1_+_207039153
|
4.020
|
NM_018724
|
IL20
|
interleukin 20
|
|
chr19_-_52227220
|
3.941
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr6_+_43211221
|
3.877
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr5_-_95768660
|
3.853
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr10_+_23983674
|
3.807
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr5_-_132948195
|
3.727
|
|
FSTL4
|
follistatin-like 4
|
|
chr17_-_8059637
|
3.718
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr5_+_137801178
|
3.639
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr2_-_157189040
|
3.523
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr19_-_46088021
|
3.504
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr5_+_159343739
|
3.367
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr16_+_71879858
|
3.362
|
|
|
|
|
chr2_+_11052062
|
3.360
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr19_+_18723666
|
3.304
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr10_-_73848766
|
3.303
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr12_-_13256572
|
3.188
|
NM_001080554
NM_001080555
NM_001206842
|
GSG1
|
germ cell associated 1
|
|
chr12_-_123380608
|
3.143
|
|
|
|
|
chr22_+_31518923
|
3.139
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr5_+_145317297
|
3.131
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr19_-_5340700
|
3.129
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr17_+_42385780
|
3.114
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr5_-_57755787
|
3.095
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr19_-_9695166
|
3.083
|
NM_001008727
|
ZNF121
|
zinc finger protein 121
|
|
chr17_+_27071008
|
3.076
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr7_+_44646220
|
3.075
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr19_+_57901330
|
3.063
|
|
ZNF548
ZNF17
|
zinc finger protein 548
zinc finger protein 17
|
|
chr17_+_20224457
|
3.045
|
|
CCDC144C
|
coiled-coil domain containing 144C
|
|
chr2_+_28615695
|
3.044
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr3_-_156877882
|
3.040
|
|
CCNL1
|
cyclin L1
|
|
chr9_+_130860816
|
3.033
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr11_-_119252364
|
3.020
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr20_+_43343884
|
3.007
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr6_-_7389941
|
2.987
|
NM_001170692
NM_001170693
NM_205864
|
CAGE1
|
cancer antigen 1
|
|
chr9_-_34397786
|
2.964
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr11_-_2193015
|
2.963
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr7_+_44646189
|
2.940
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr2_+_74781511
|
2.938
|
NM_001381
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr10_-_99446899
|
2.934
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr19_-_4902878
|
2.900
|
NM_001080523
|
ARRDC5
|
arrestin domain containing 5
|
|
chr7_+_44646168
|
2.879
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr9_+_132251323
|
2.869
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr19_-_9731883
|
2.868
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr10_-_128209996
|
2.852
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr10_+_101542460
|
2.822
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr3_+_89156804
|
2.814
|
|
EPHA3
|
EPH receptor A3
|
|
chr16_+_71879893
|
2.791
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr2_+_28615771
|
2.771
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr9_+_130830595
|
2.751
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr14_-_77494621
|
2.748
|
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr7_+_44646197
|
2.718
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr17_+_53342320
|
2.716
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr9_+_130830447
|
2.682
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr21_-_35899046
|
2.670
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr19_-_2702738
|
2.611
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr19_-_48018277
|
2.600
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr12_+_86268072
|
2.574
|
NM_006183
|
NTS
|
neurotensin
|
|
chr17_+_71161110
|
2.558
|
NM_001050
|
SSTR2
|
somatostatin receptor 2
|
|
chr12_-_12715244
|
2.547
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr10_-_73848530
|
2.518
|
NM_001244950
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr11_+_24518515
|
2.516
|
NM_001009909
NM_001252008
NM_001252010
|
LUZP2
|
leucine zipper protein 2
|
|
chr19_-_49540072
|
2.511
|
NM_033377
|
CGB1
CGB2
|
chorionic gonadotropin, beta polypeptide 1
chorionic gonadotropin, beta polypeptide 2
|
|
chr7_+_128312345
|
2.504
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr20_+_33292117
|
2.475
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr1_+_152974222
|
2.473
|
NM_001097589
NM_005416
|
SPRR3
|
small proline-rich protein 3
|
|
chr10_-_106093662
|
2.470
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr17_+_48046561
|
2.459
|
NM_138281
|
DLX4
|
distal-less homeobox 4
|
|
chr9_-_131644176
|
2.443
|
NM_001122671
NM_001122672
NM_004059
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic
|
|
chr19_-_48018163
|
2.442
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr9_-_33402505
|
2.436
|
NM_001170
|
AQP7
|
aquaporin 7
|
|
chr13_+_46276445
|
2.420
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr14_+_96722546
|
2.418
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr10_-_131762374
|
2.373
|
|
EBF3
|
early B-cell factor 3
|
|
chr6_-_26032287
|
2.352
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr2_-_27531113
|
2.327
|
NM_003353
|
UCN
|
urocortin
|
|
chr17_+_53342356
|
2.310
|
|
HLF
|
hepatic leukemia factor
|
|
chrX_+_83116153
|
2.309
|
NM_021118
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1
|
|
chr19_+_45504686
|
2.304
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr15_-_72612497
|
2.296
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr18_-_77439739
|
2.293
|
|
FLJ25715
|
uncharacterized protein FLJ25715
|
|
chr20_+_55099784
|
2.291
|
NM_001012971
|
FAM209A
FAM209B
|
family with sequence similarity 209, member A
family with sequence similarity 209, member B
|
|
chr10_-_106098161
|
2.274
|
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr1_+_45266035
|
2.261
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr1_-_161039745
|
2.252
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr11_-_119252267
|
2.243
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr7_+_44646180
|
2.238
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr14_-_69259654
|
2.236
|
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr19_+_54960380
|
2.234
|
|
LENG8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr2_-_134326030
|
2.220
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr20_+_17207596
|
2.217
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr19_-_2702662
|
2.209
|
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr5_-_138211054
|
2.206
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr1_-_76076762
|
2.169
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr3_+_89156673
|
2.162
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr22_+_50528307
|
2.161
|
NM_001164104
NM_018995
|
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse)
|
|
chr19_-_46088084
|
2.155
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr9_-_73028278
|
2.154
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr15_+_31619043
|
2.150
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr18_+_13218777
|
2.149
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr9_+_132251403
|
2.147
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr6_-_166401526
|
2.136
|
|
LINC00473
|
long intergenic non-protein coding RNA 473
|
|
chr20_+_32250089
|
2.135
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr4_+_145567147
|
2.120
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr22_+_21771661
|
2.099
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr2_+_211458078
|
2.085
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr7_+_73245192
|
2.084
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr14_+_93389491
|
2.075
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr8_-_29207795
|
2.057
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr20_+_10199455
|
2.018
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr20_-_39317875
|
2.018
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr19_-_46088074
|
2.011
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr17_-_77813049
|
2.007
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr1_+_9599520
|
2.005
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr6_-_27782517
|
2.005
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr16_+_86544104
|
2.004
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr1_+_9599546
|
2.002
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr2_+_28615668
|
2.001
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr12_+_104359557
|
1.999
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr5_-_112770537
|
1.991
|
NM_032028
|
TSSK1B
|
testis-specific serine kinase 1B
|
|
chr2_-_157186681
|
1.988
|
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr15_+_78556901
|
1.952
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr3_-_62359189
|
1.952
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr8_+_126442557
|
1.948
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr19_+_49109999
|
1.948
|
NM_133498
|
SPACA4
|
sperm acrosome associated 4
|
|
chr14_-_69260630
|
1.925
|
NM_001244698
NM_004926
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr16_+_69139466
|
1.925
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chrX_+_110339374
|
1.923
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr7_+_44646120
|
1.893
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr9_+_105757592
|
1.890
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr8_-_95274520
|
1.879
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr19_-_55628780
|
1.877
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr19_+_54960064
|
1.874
|
NM_052925
|
LENG8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr15_-_72612127
|
1.871
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_-_72612469
|
1.859
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_-_217113007
|
1.852
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr17_-_8661876
|
1.833
|
NM_001128076
|
SPDYE4
|
speedy homolog E4 (Xenopus laevis)
|
|
chr1_+_45265896
|
1.830
|
|
PLK3
|
polo-like kinase 3
|
|
chr1_-_161039413
|
1.813
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr8_-_95274540
|
1.810
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr17_+_48585744
|
1.807
|
NM_032133
|
MYCBPAP
|
MYCBP associated protein
|
|
chr1_+_45266048
|
1.805
|
|
PLK3
|
polo-like kinase 3
|
|
chr20_-_44600939
|
1.799
|
|
ZNF335
|
zinc finger protein 335
|
|
chr19_-_57352055
|
1.788
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr11_+_65658101
|
1.787
|
|
CCDC85B
|
coiled-coil domain containing 85B
|
|
chr1_-_161039703
|
1.786
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr17_-_40828441
|
1.784
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr9_-_19033183
|
1.779
|
NM_153707
|
FAM154A
|
family with sequence similarity 154, member A
|
|
chr2_-_28113954
|
1.777
|
|
LOC100302650
|
uncharacterized LOC100302650
|
|
chrX_+_9431314
|
1.771
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_-_161039649
|
1.763
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr8_-_101322245
|
1.750
|
NM_183419
|
RNF19A
|
ring finger protein 19A
|