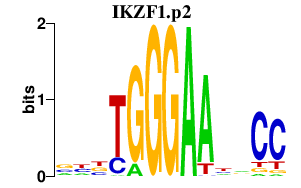
|
chr15_-_42448769
|
15.702
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr6_+_126070813
|
12.032
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr6_+_126070895
|
9.206
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr4_-_57522059
|
6.320
|
|
HOPX
|
HOP homeobox
|
|
chr1_-_109849613
|
5.710
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr6_+_126070724
|
5.702
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr6_+_126070738
|
5.575
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr19_-_36019194
|
5.032
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr10_-_5541401
|
4.966
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr17_+_38474513
|
4.653
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr17_+_38474544
|
4.434
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr19_-_51456159
|
4.002
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr14_+_79745653
|
3.876
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr9_+_138453599
|
3.842
|
NM_001018049
NM_002571
|
PAEP
|
progestagen-associated endometrial protein
|
|
chr17_-_3461030
|
3.238
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chrX_+_115567820
|
3.190
|
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr12_+_41086189
|
3.161
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chrX_+_115567746
|
3.119
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr9_-_21994414
|
3.087
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr9_+_17578952
|
3.034
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr9_-_21994329
|
3.005
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr19_+_42259397
|
3.004
|
NM_002483
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen)
|
|
chr11_-_2018703
|
2.986
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr2_+_17721983
|
2.974
|
|
VSNL1
|
visinin-like 1
|
|
chr7_+_148036528
|
2.763
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr2_+_17721806
|
2.750
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr22_+_37257004
|
2.743
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr13_-_36429798
|
2.690
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr15_+_75118950
|
2.647
|
NM_001030005
|
CPLX3
|
complexin 3
|
|
chr12_-_58160823
|
2.628
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr19_+_496453
|
2.624
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr9_-_21994379
|
2.578
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr4_-_57547845
|
2.515
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr7_-_148581403
|
2.491
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_+_160336856
|
2.445
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr17_+_38474472
|
2.423
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr7_+_47694841
|
2.384
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr7_-_148581379
|
2.379
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_+_15250578
|
2.370
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr22_-_19512859
|
2.363
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr4_+_159690208
|
2.351
|
|
FNIP2
|
folliculin interacting protein 2
|
|
chr12_-_45307710
|
2.333
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr19_+_46801632
|
2.248
|
NM_022462
NM_152796
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr3_+_127634317
|
2.222
|
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr2_-_224467095
|
2.179
|
|
SCG2
|
secretogranin II
|
|
chr20_+_56136136
|
2.091
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr17_+_38465422
|
2.089
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr1_+_113263188
|
2.089
|
NM_001004440
NM_182759
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3
|
|
chr2_+_102608305
|
2.067
|
NM_004633
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr7_-_148581377
|
2.036
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_-_148581297
|
2.007
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr8_-_70747200
|
1.994
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr9_-_33447527
|
1.975
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr1_-_152386727
|
1.974
|
NM_016190
|
CRNN
|
cornulin
|
|
chr16_-_66952729
|
1.971
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr2_-_224467142
|
1.943
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr2_-_224467049
|
1.935
|
|
SCG2
|
secretogranin II
|
|
chr7_+_23286386
|
1.922
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_-_148581359
|
1.921
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_+_33945111
|
1.918
|
NM_133468
|
BMPER
|
BMP binding endothelial regulator
|
|
chr4_+_81256873
|
1.909
|
NM_001206997
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chr17_-_39646115
|
1.895
|
NM_003771
|
KRT36
|
keratin 36
|
|
chr17_+_38119225
|
1.893
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr19_-_10121041
|
1.891
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr5_-_95018659
|
1.889
|
NM_031952
|
SPATA9
|
spermatogenesis associated 9
|
|
chr2_+_16080639
|
1.887
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr19_+_49109999
|
1.881
|
NM_133498
|
SPACA4
|
sperm acrosome associated 4
|
|
chr10_+_85933553
|
1.875
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr1_+_26856240
|
1.869
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr12_-_322612
|
1.860
|
NM_003044
NM_001122847
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12
|
|
chr10_-_50396380
|
1.857
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr11_+_3666357
|
1.845
|
NM_004314
|
ART1
|
ADP-ribosyltransferase 1
|
|
chr19_-_6333561
|
1.834
|
NM_133492
|
ACER1
|
alkaline ceramidase 1
|
|
chr11_-_35441501
|
1.829
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_-_160039946
|
1.810
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr16_+_29690464
|
1.808
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr16_+_71879893
|
1.798
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr11_+_72980938
|
1.793
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr1_+_26856252
|
1.788
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr7_+_23286384
|
1.787
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286420
|
1.777
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_176875325
|
1.773
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr17_-_32483550
|
1.772
|
NM_001094
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr2_+_121103670
|
1.739
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr7_+_23286390
|
1.733
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr17_-_72588369
|
1.725
|
NM_001115152
|
CD300LD
|
CD300 molecule-like family member d
|
|
chr14_-_48144156
|
1.724
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr19_-_42498320
|
1.715
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr15_-_93198602
|
1.710
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr19_-_51504799
|
1.709
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr16_-_3068161
|
1.706
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr2_+_16080559
|
1.696
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr5_-_156772672
|
1.684
|
NM_001001343
|
FNDC9
|
fibronectin type III domain containing 9
|
|
chr19_+_19322763
|
1.681
|
NM_004386
|
NCAN
|
neurocan
|
|
chr13_+_37005966
|
1.677
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr11_-_35441099
|
1.658
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr19_+_46800296
|
1.657
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr2_+_3642421
|
1.646
|
NM_024027
NM_199235
|
COLEC11
|
collectin sub-family member 11
|
|
chr7_+_150745604
|
1.641
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr2_-_109605616
|
1.634
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr17_+_25799024
|
1.622
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr19_+_15751706
|
1.621
|
NM_000896
NM_001199208
NM_001199209
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr15_-_73661141
|
1.617
|
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr20_-_23030141
|
1.603
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr21_-_31538970
|
1.603
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr11_-_10715160
|
1.602
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr8_-_86789305
|
1.592
|
NM_172239
|
REXO1L1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1
|
|
chr1_-_203154304
|
1.581
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chrX_-_118284541
|
1.580
|
NM_020721
|
KIAA1210
|
KIAA1210
|
|
chr8_+_48873533
|
1.577
|
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chr11_-_35440514
|
1.576
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr8_-_98290171
|
1.574
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr1_+_26856259
|
1.547
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr19_-_42498221
|
1.543
|
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr2_+_27371858
|
1.536
|
NM_175769
|
TCF23
|
transcription factor 23
|
|
chr19_-_10676689
|
1.521
|
NM_023008
|
KRI1
|
KRI1 homolog (S. cerevisiae)
|
|
chr3_+_124103551
|
1.510
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr2_-_193059249
|
1.496
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr10_-_102890902
|
1.495
|
NM_001085398
|
TLX1NB
|
TLX1 neighbor
|
|
chr22_-_30162990
|
1.489
|
|
|
|
|
chr5_-_160112266
|
1.486
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr10_+_102891060
|
1.479
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chr1_-_40105341
|
1.478
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr10_+_115511212
|
1.475
|
NM_001193434
NM_001193435
NM_024889
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr1_-_153321021
|
1.474
|
NM_020393
|
PGLYRP4
|
peptidoglycan recognition protein 4
|
|
chr10_-_131762017
|
1.471
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr12_-_57328044
|
1.470
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr6_+_31082526
|
1.466
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr3_-_170303774
|
1.464
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr5_-_131563481
|
1.460
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr2_+_179059071
|
1.458
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr19_-_55549612
|
1.456
|
NM_001083899
NM_016363
|
GP6
|
glycoprotein VI (platelet)
|
|
chr19_-_10676646
|
1.455
|
|
KRI1
|
KRI1 homolog (S. cerevisiae)
|
|
chr8_-_145669796
|
1.452
|
NM_013432
|
TONSL
|
tonsoku-like, DNA repair protein
|
|
chr1_-_202130677
|
1.452
|
NM_001199797
NM_002832
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr12_-_43945723
|
1.451
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr2_-_39187477
|
1.449
|
|
LOC375196
|
uncharacterized LOC375196
|
|
chr1_-_27952591
|
1.445
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr1_-_36235422
|
1.442
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr12_+_113495661
|
1.431
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr17_+_38024416
|
1.419
|
NM_198844
NM_199321
|
ZPBP2
|
zona pellucida binding protein 2
|
|
chr4_+_7194317
|
1.416
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr19_-_45926759
|
1.411
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr11_-_111250156
|
1.408
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr12_-_58159386
|
1.403
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr6_-_90024936
|
1.402
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr19_-_19384022
|
1.397
|
NM_001001524
|
TM6SF2
|
transmembrane 6 superfamily member 2
|
|
chr4_-_40631846
|
1.396
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_+_151009028
|
1.389
|
NM_001159642
NM_138278
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr4_-_74864296
|
1.386
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr2_-_100722328
|
1.384
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr11_-_111249902
|
1.379
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr12_+_112924284
|
1.371
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr1_-_40105296
|
1.370
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr5_+_175298538
|
1.367
|
|
CPLX2
|
complexin 2
|
|
chr7_-_126892304
|
1.365
|
NM_001127323
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chrX_-_112084006
|
1.360
|
NM_133265
|
AMOT
|
angiomotin
|
|
chrX_+_55246790
|
1.354
|
NM_130467
NM_001013435
|
PAGE5
|
P antigen family, member 5 (prostate associated)
|
|
chr19_-_45926819
|
1.349
|
NM_202001
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr10_-_6104113
|
1.348
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr9_+_87283453
|
1.348
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr8_-_98290072
|
1.343
|
|
TSPYL5
|
TSPY-like 5
|
|
chr3_-_112013073
|
1.339
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr12_+_48592139
|
1.337
|
|
DKFZP779L1853
|
uncharacterized LOC643162
|
|
chr17_-_7493389
|
1.336
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr18_+_48086402
|
1.335
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr5_+_75379238
|
1.326
|
NM_014979
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
|
chr4_-_155533850
|
1.324
|
|
FGG
|
fibrinogen gamma chain
|
|
chr14_+_24521099
|
1.324
|
NM_138360
|
LRRC16B
|
leucine rich repeat containing 16B
|
|
chr16_+_23690234
|
1.323
|
|
PLK1
|
polo-like kinase 1
|
|
chr11_+_1860232
|
1.318
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr4_+_159690176
|
1.317
|
NM_020840
|
FNIP2
|
folliculin interacting protein 2
|
|
chr6_-_94129059
|
1.311
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr4_-_72671236
|
1.308
|
NM_001204306
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr1_+_176432306
|
1.303
|
NM_020318
NM_021936
|
PAPPA2
|
pappalysin 2
|
|
chr17_+_7591666
|
1.296
|
NM_001143992
NM_018081
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr4_-_155412411
|
1.295
|
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr20_-_45179212
|
1.288
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr11_-_10715297
|
1.271
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr6_+_54173202
|
1.270
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr19_+_38085730
|
1.268
|
NM_001172225
NM_001172226
|
ZNF540
|
zinc finger protein 540
|
|
chr11_+_64126616
|
1.267
|
NM_001006944
NM_003942
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4
|
|
chr16_+_71392615
|
1.263
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr19_+_50472856
|
1.258
|
|
SIGLEC16
|
sialic acid binding Ig-like lectin 16 (gene/pseudogene)
|
|
chr20_-_35201677
|
1.256
|
|
|
|
|
chr19_+_46806855
|
1.256
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr10_+_123872440
|
1.254
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr15_+_59730272
|
1.252
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr19_-_45926631
|
1.251
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr1_-_182030749
|
1.249
|
NM_001009992
|
ZNF648
|
zinc finger protein 648
|
|
chr16_+_23690212
|
1.241
|
|
PLK1
|
polo-like kinase 1
|
|
chr8_-_23261632
|
1.241
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_-_155533800
|
1.239
|
|
FGG
|
fibrinogen gamma chain
|
|
chr10_+_115939018
|
1.239
|
NM_198795
|
TDRD1
|
tudor domain containing 1
|
|
chr8_-_86575725
|
1.228
|
NM_172239
|
REXO1L1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1
|
|
chrX_+_102470003
|
1.227
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr4_-_155533882
|
1.224
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_68864341
|
1.223
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr21_+_27012067
|
1.218
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr13_-_38443859
|
1.217
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|