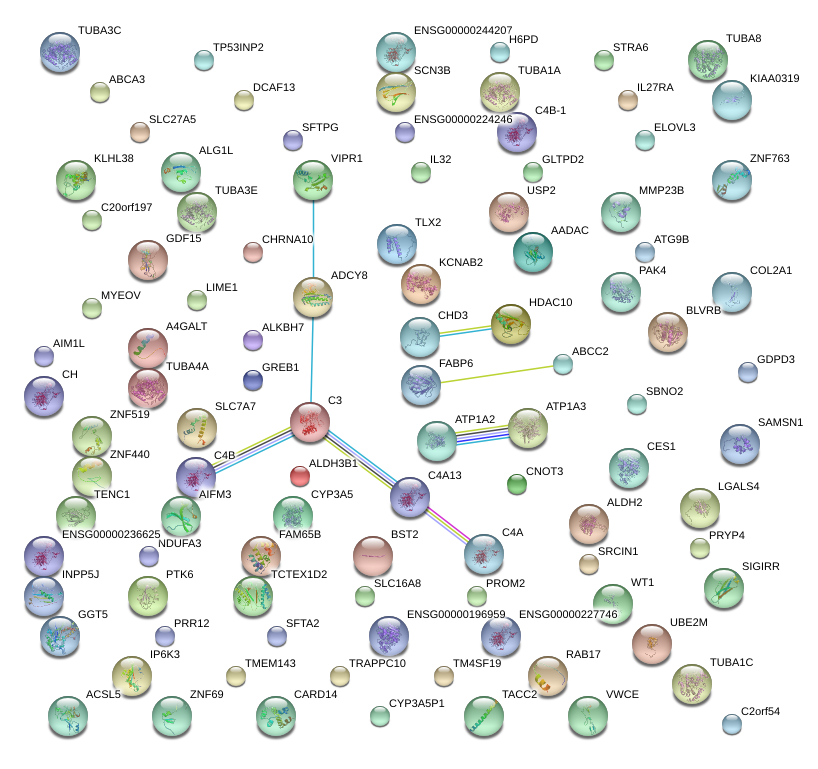
|
chr5_+_159656436
|
10.730
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr17_+_78152280
|
9.849
|
NM_024110
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr1_-_29309092
|
9.585
|
|
|
|
|
chr10_+_101542554
|
9.421
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr22_+_21321446
|
9.333
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr10_+_101542460
|
9.287
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr19_+_18496966
|
9.160
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr2_+_11680079
|
8.948
|
NM_148903
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr19_+_45116921
|
8.944
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr2_+_11674241
|
8.651
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr20_-_62168663
|
8.145
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr11_-_61062534
|
7.992
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr10_+_103986142
|
7.711
|
NM_152310
|
ELOVL3
|
ELOVL fatty acid elongase 3
|
|
chr19_+_1776117
|
7.644
|
|
|
|
|
chr5_+_133842276
|
7.497
|
|
|
|
|
chr15_-_74500394
|
7.387
|
NM_001199041
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr3_-_196065191
|
7.184
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr19_-_17516383
|
7.149
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr6_-_24877453
|
7.115
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr7_-_99277518
|
6.961
|
NM_000777
NM_001190484
|
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5
|
|
chr17_+_7812703
|
6.917
|
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr16_-_30124829
|
6.744
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr2_+_95940220
|
6.722
|
|
PROM2
|
prominin 2
|
|
chr16_+_3115312
|
6.620
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr19_-_59023323
|
6.555
|
NM_012254
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5
|
|
chr19_+_11998609
|
6.526
|
NM_021915
|
ZNF69
|
zinc finger protein 69
|
|
chr16_-_55867016
|
6.475
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr19_-_48867149
|
6.339
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr16_-_55866972
|
6.224
|
|
CES1
|
carboxylesterase 1
|
|
chr6_-_33714681
|
5.910
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr5_-_42811959
|
5.845
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr3_-_196065243
|
5.701
|
NM_001204897
NM_001204898
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr19_-_39402797
|
5.567
|
NM_001243212
|
LOC643669
|
uncharacterized LOC643669
|
|
chr20_+_33299315
|
5.549
|
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr1_+_1567556
|
5.440
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr11_-_123525300
|
5.409
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr19_-_39303520
|
5.381
|
|
LGALS4
|
lectin, galactoside-binding, soluble, 4
|
|
chr19_-_59069747
|
5.346
|
|
UBE2M
|
ubiquitin-conjugating enzyme E2M
|
|
chr2_+_95940195
|
5.297
|
NM_001165977
NM_001165978
NM_144707
|
PROM2
|
prominin 2
|
|
chr18_-_14132426
|
5.262
|
NM_145287
|
ZNF519
|
zinc finger protein 519
|
|
chr8_-_124665165
|
5.258
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr10_+_114133838
|
5.245
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_+_6052357
|
5.238
|
NM_001199861
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr14_-_23284617
|
5.236
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr11_-_61062697
|
5.219
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr1_+_160085519
|
5.214
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr2_-_241831385
|
5.200
|
NM_024861
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr20_+_58630965
|
5.183
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr2_-_130956011
|
5.179
|
NM_207312
|
TUBA3E
|
tubulin, alpha 3e
|
|
chr7_-_150721551
|
5.051
|
NM_173681
|
ATG9B
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chr3_+_151531860
|
4.972
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr21_-_15918635
|
4.926
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr19_+_54606141
|
4.848
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr17_+_4692226
|
4.843
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr21_+_45432202
|
4.780
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr1_+_9299902
|
4.748
|
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr10_+_114133775
|
4.740
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr22_-_24640850
|
4.701
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr17_+_77893155
|
4.684
|
|
|
|
|
chr3_+_42530790
|
4.660
|
NM_001251882
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr3_-_125655881
|
4.628
|
NM_001015050
NM_001195223
|
ALG1L
|
asparagine-linked glycosylation 1-like
|
|
chr19_+_50094911
|
4.608
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr6_-_24645955
|
4.604
|
NM_001168375
NM_001168376
NM_001168374
NM_001168377
NM_014809
|
KIAA0319
|
KIAA0319
|
|
chr19_-_6720585
|
4.500
|
NM_000064
|
C3
|
complement component 3
|
|
chr1_-_26680620
|
4.472
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr11_+_69061599
|
4.373
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr10_+_123922940
|
4.294
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_+_74741565
|
4.286
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr19_+_54641420
|
4.266
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr19_-_40971623
|
4.245
|
NM_000713
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH))
|
|
chr17_-_36729054
|
4.223
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr22_+_31518923
|
4.222
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr19_+_54641475
|
4.218
|
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr6_+_31982538
|
4.217
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr22_-_50689460
|
4.198
|
|
HDAC10
|
histone deacetylase 10
|
|
chr19_+_14142261
|
4.188
|
NM_004843
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr15_-_74494780
|
4.176
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr19_+_39616384
|
4.157
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr11_+_67777795
|
4.140
|
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr11_-_119234628
|
4.082
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr19_+_6372805
|
4.036
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr12_-_48398284
|
4.029
|
NM_001844
NM_033150
|
COL2A1
|
collagen, type II, alpha 1
|
|
chr6_-_30899926
|
4.014
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr11_-_32457059
|
3.912
|
NM_000378
NM_024424
NM_024426
|
WT1
|
Wilms tumor 1
|
|
chr11_+_67777788
|
3.898
|
NM_000694
NM_001030010
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr12_+_53443834
|
3.892
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr22_-_43089856
|
3.887
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr19_-_1174263
|
3.885
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr2_-_238499317
|
3.827
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr22_-_38479148
|
3.824
|
NM_013356
|
SLC16A8
|
solute carrier family 16, member 8 (monocarboxylic acid transporter 3)
|
|
chr11_-_414975
|
3.813
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr11_-_3692544
|
3.812
|
NM_020402
|
CHRNA10
|
cholinergic receptor, nicotinic, alpha 10
|
|
chr17_-_73840422
|
3.765
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr2_+_11886739
|
3.743
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr12_-_54779482
|
3.736
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr11_+_66742747
|
3.711
|
NM_001136485
|
C11orf86
|
chromosome 11 open reading frame 86
|
|
chr19_+_6372443
|
3.710
|
NM_032306
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr15_-_74494851
|
3.694
|
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr17_-_73840495
|
3.694
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr2_+_219187975
|
3.646
|
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr19_-_58892382
|
3.642
|
NM_001129730
NM_138466
|
ZNF837
|
zinc finger protein 837
|
|
chr12_-_8043743
|
3.630
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr17_+_72439187
|
3.591
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr19_-_1174201
|
3.590
|
|
|
|
|
chr14_-_23285011
|
3.587
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr4_+_4543718
|
3.580
|
|
LOC100507266
|
uncharacterized LOC100507266
|
|
chr1_+_20915440
|
3.572
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr15_+_85427891
|
3.570
|
NM_004213
NM_201651
|
SLC28A1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1
|
|
chr17_-_8198605
|
3.568
|
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr11_+_1861431
|
3.566
|
NM_001145841
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr17_-_19651586
|
3.555
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr19_+_44507076
|
3.553
|
NM_006300
|
ZNF230
|
zinc finger protein 230
|
|
chr19_-_55580896
|
3.542
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr19_-_38806546
|
3.542
|
NM_001039672
NM_001039673
NM_001145463
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr19_-_41255827
|
3.503
|
NM_198476
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr22_-_50689766
|
3.482
|
NM_001159286
NM_032019
|
HDAC10
|
histone deacetylase 10
|
|
chr19_+_46651499
|
3.482
|
NM_001135113
|
IGFL2
|
IGF-like family member 2
|
|
chr22_-_50689685
|
3.467
|
|
HDAC10
|
histone deacetylase 10
|
|
chrX_-_15511358
|
3.452
|
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr6_-_45983279
|
3.448
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr20_+_31367637
|
3.446
|
NM_175850
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr9_-_95896498
|
3.442
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr19_+_6372720
|
3.441
|
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chrX_-_15511710
|
3.422
|
NM_001018109
NM_003662
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr11_+_64053581
|
3.385
|
NM_001170880
NM_020155
|
GPR137
|
G protein-coupled receptor 137
|
|
chr19_+_49458129
|
3.383
|
|
BAX
|
BCL2-associated X protein
|
|
chr11_-_65325391
|
3.372
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr19_+_49458151
|
3.355
|
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_49128208
|
3.354
|
NM_001204160
|
SPHK2
|
sphingosine kinase 2
|
|
chr5_+_179220985
|
3.354
|
NM_145867
|
LTC4S
|
leukotriene C4 synthase
|
|
chr19_-_1174257
|
3.326
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr3_-_62359189
|
3.325
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr4_-_53617781
|
3.325
|
NM_001242690
NM_024534
|
ERVMER34-1
|
endogenous retrovirus group MER34, member 1
|
|
chr19_+_49458031
|
3.322
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr13_+_31287614
|
3.314
|
NM_001204406
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr3_-_48671278
|
3.312
|
NM_001040454
|
SLC26A6
|
solute carrier family 26, member 6
|
|
chr19_-_49622278
|
3.307
|
NM_018111
|
C19orf73
|
chromosome 19 open reading frame 73
|
|
chr19_+_45251933
|
3.305
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr15_-_83349134
|
3.300
|
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr3_+_42544131
|
3.288
|
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr1_+_61547625
|
3.283
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr6_+_138188352
|
3.278
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_-_155271186
|
3.273
|
NM_000298
|
PKLR
|
pyruvate kinase, liver and RBC
|
|
chr2_+_211458078
|
3.260
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr16_-_70719693
|
3.209
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr3_+_42544094
|
3.208
|
NM_001251883
NM_001251884
NM_001251885
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr6_+_31913708
|
3.208
|
NM_001710
|
CFB
|
complement factor B
|
|
chr22_-_24059502
|
3.208
|
|
GUSBP11
|
glucuronidase, beta pseudogene 11
|
|
chr2_+_211421322
|
3.183
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr10_+_70847857
|
3.181
|
|
SRGN
|
serglycin
|
|
chr16_+_56225250
|
3.177
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr2_-_132559233
|
3.172
|
NM_214461
|
C2orf27B
|
chromosome 2 open reading frame 27B
|
|
chr22_+_25003610
|
3.166
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr1_+_26348265
|
3.142
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr19_-_55628780
|
3.131
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C
|
|
chr14_-_65409445
|
3.124
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr17_+_18281078
|
3.120
|
NM_001145127
|
EVPLL
|
envoplakin-like
|
|
chr10_-_104211244
|
3.109
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr17_+_7905911
|
3.103
|
NM_000180
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific)
|
|
chr17_-_8027396
|
3.103
|
NM_001165967
NM_032580
|
HES7
|
hairy and enhancer of split 7 (Drosophila)
|
|
chr22_-_24059436
|
3.094
|
|
|
|
|
chr11_-_32452358
|
3.080
|
NM_001198551
NM_001198552
|
WT1
|
Wilms tumor 1
|
|
chr14_+_39735500
|
3.072
|
NM_001247988
NM_203356
|
CTAGE5
|
CTAGE family, member 5
|
|
chr19_+_46498338
|
3.058
|
NM_001080402
|
CCDC61
|
coiled-coil domain containing 61
|
|
chr9_+_35906188
|
3.051
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr17_-_1553099
|
3.043
|
NM_031430
|
RILP
|
Rab interacting lysosomal protein
|
|
chr11_-_6462129
|
3.041
|
NM_000613
|
HPX
|
hemopexin
|
|
chr19_-_47987410
|
3.031
|
|
KPTN
|
kaptin (actin binding protein)
|
|
chr11_-_1593149
|
3.030
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr11_-_116663106
|
3.028
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr16_+_66995099
|
3.015
|
NM_001185177
NM_024922
|
CES3
|
carboxylesterase 3
|
|
chr19_-_18385166
|
3.014
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr12_+_113659216
|
3.009
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr1_-_92949500
|
3.002
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr17_+_48050129
|
2.999
|
NM_001934
|
DLX4
|
distal-less homeobox 4
|
|
chr3_+_194406614
|
2.999
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr7_+_76751933
|
2.995
|
NM_020879
|
CCDC146
|
coiled-coil domain containing 146
|
|
chr10_-_91295310
|
2.995
|
NM_213606
|
SLC16A12
|
solute carrier family 16, member 12 (monocarboxylic acid transporter 12)
|
|
chr2_+_11682836
|
2.987
|
NM_033090
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr1_-_155881063
|
2.981
|
|
RIT1
|
Ras-like without CAAX 1
|
|
chr14_+_75745480
|
2.972
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr3_-_49459870
|
2.948
|
|
AMT
|
aminomethyltransferase
|
|
chr19_-_19281062
|
2.942
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr19_+_45754505
|
2.934
|
NM_001199867
NM_031417
|
MARK4
|
MAP/microtubule affinity-regulating kinase 4
|
|
chr16_+_3115639
|
2.923
|
|
IL32
|
interleukin 32
|
|
chr2_+_234601511
|
2.917
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr1_+_26146446
|
2.915
|
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr12_-_6484904
|
2.901
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr2_+_11682787
|
2.901
|
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr19_-_42498320
|
2.895
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr14_+_103592663
|
2.872
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr17_+_48049678
|
2.863
|
|
DLX4
|
distal-less homeobox 4
|
|
chr6_+_31926788
|
2.855
|
|
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae)
|
|
chr15_-_74502045
|
2.853
|
NM_001199040
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr1_+_44457424
|
2.844
|
|
CCDC24
|
coiled-coil domain containing 24
|
|
chr1_+_222988324
|
2.834
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr22_-_51021255
|
2.825
|
|
CHKB
|
choline kinase beta
|
|
chr19_-_38806381
|
2.822
|
NM_001039671
NM_001145461
NM_001145462
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr22_-_42526792
|
2.815
|
|
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6
|
|
chr19_-_1111972
|
2.812
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|