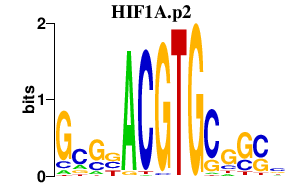
|
chr2_+_113403433
|
7.929
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr6_+_7726268
|
7.271
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr7_+_156931609
|
6.841
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr7_+_156931631
|
6.729
|
NM_014671
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr7_+_156931663
|
6.404
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr5_+_176873795
|
6.256
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr2_-_75062214
|
5.766
|
|
|
|
|
chr3_-_183543295
|
5.628
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr11_+_12132063
|
5.510
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr18_+_43913906
|
5.357
|
|
RNF165
|
ring finger protein 165
|
|
chr17_-_3599298
|
5.262
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr13_+_41363754
|
5.120
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr3_-_16555128
|
4.971
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr13_+_41363546
|
4.911
|
NM_014252
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr13_+_41363642
|
4.908
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr7_+_128095881
|
4.554
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chr1_-_182573393
|
4.483
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr3_-_127541160
|
4.378
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr3_-_16554992
|
4.358
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr4_-_4291817
|
4.318
|
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr10_+_90639900
|
4.196
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr1_-_212873104
|
4.147
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr4_-_4291731
|
4.144
|
NM_001145725
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr16_+_222845
|
4.131
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr11_+_18416111
|
4.014
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr2_-_227663454
|
3.997
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr16_+_226678
|
3.982
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr3_-_145879222
|
3.981
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr11_+_18416106
|
3.952
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr3_-_127541975
|
3.889
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr17_-_3599359
|
3.882
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr13_-_20806533
|
3.816
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr8_-_75233461
|
3.728
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr3_-_52090090
|
3.661
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chrX_+_16804531
|
3.640
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr3_-_170626381
|
3.628
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr6_+_43738757
|
3.617
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_+_60609428
|
3.543
|
NM_024098
|
CCDC86
|
coiled-coil domain containing 86
|
|
chr8_-_145515014
|
3.527
|
|
BOP1
|
block of proliferation 1
|
|
chr11_+_18415917
|
3.478
|
NM_001135239
NM_001165415
NM_001165416
NM_005566
|
LDHA
|
lactate dehydrogenase A
|
|
chr17_-_41622727
|
3.445
|
|
ETV4
|
ets variant 4
|
|
chr3_-_170626425
|
3.389
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_+_52719935
|
3.357
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr15_-_42264749
|
3.305
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr6_+_151646634
|
3.275
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr15_-_42264687
|
3.272
|
|
EHD4
|
EH-domain containing 4
|
|
chr22_+_45098046
|
3.246
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr9_-_140196702
|
3.216
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chrX_+_68725077
|
3.211
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr1_+_193091057
|
3.190
|
NM_024529
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr1_-_41328002
|
3.174
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr8_+_74888376
|
3.165
|
NM_001040613
NM_017866
|
TMEM70
|
transmembrane protein 70
|
|
chr17_-_73874056
|
3.140
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr17_-_36831156
|
3.006
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr6_+_10695155
|
2.985
|
NM_017906
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr13_+_100633977
|
2.957
|
NM_007129
|
ZIC2
|
Zic family member 2
|
|
chr2_+_68384986
|
2.931
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr7_-_105162551
|
2.927
|
NM_019042
|
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae)
|
|
chr12_+_122150638
|
2.923
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr1_-_6526150
|
2.908
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chr10_-_33623299
|
2.892
|
|
NRP1
|
neuropilin 1
|
|
chr3_-_145878936
|
2.850
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr1_+_193091220
|
2.836
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr17_-_41623246
|
2.833
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr1_+_65613211
|
2.818
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr8_-_13133956
|
2.817
|
|
|
|
|
chr22_-_30987819
|
2.797
|
|
PES1
|
pescadillo homolog 1, containing BRCT domain (zebrafish)
|
|
chr1_+_193091165
|
2.786
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr10_+_3110818
|
2.776
|
NM_001242339
|
PFKP
|
phosphofructokinase, platelet
|
|
chr8_+_31497267
|
2.754
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr17_-_2614926
|
2.752
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr1_+_193091283
|
2.731
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr1_-_247335251
|
2.728
|
NM_001243740
NM_003431
|
ZNF124
|
zinc finger protein 124
|
|
chr19_+_18530087
|
2.728
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chrX_-_152864435
|
2.722
|
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr6_+_111303240
|
2.696
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr22_-_30987866
|
2.683
|
NM_001243225
NM_014303
|
PES1
|
pescadillo homolog 1, containing BRCT domain (zebrafish)
|
|
chr3_-_50374795
|
2.666
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr8_+_38244647
|
2.648
|
NM_001199660
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr19_+_38810540
|
2.640
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr1_+_193091196
|
2.594
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr8_-_23261632
|
2.592
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr9_-_135545327
|
2.583
|
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31
|
|
chr19_+_4909447
|
2.569
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr4_-_7069755
|
2.568
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr8_+_6566153
|
2.563
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr17_+_4402128
|
2.557
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr1_+_19578039
|
2.530
|
NM_016183
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae)
|
|
chr16_+_56966025
|
2.521
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr9_+_112542496
|
2.490
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr15_-_93198602
|
2.481
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr16_+_22825806
|
2.478
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr1_-_6557471
|
2.476
|
NM_001042663
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_+_65613971
|
2.455
|
|
AK4
|
adenylate kinase 4
|
|
chr13_+_28494136
|
2.452
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr19_+_18530211
|
2.451
|
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr17_+_45000483
|
2.448
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr7_+_72848108
|
2.447
|
NM_003508
|
FZD9
|
frizzled family receptor 9
|
|
chr13_-_41706587
|
2.442
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr3_-_50329702
|
2.436
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr12_-_50677238
|
2.426
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr8_-_23261626
|
2.423
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr17_-_13505217
|
2.412
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr17_+_80186886
|
2.410
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr20_+_48599511
|
2.408
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr10_+_101419169
|
2.400
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr17_+_40985412
|
2.396
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr16_-_2264776
|
2.395
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr3_+_149689065
|
2.392
|
|
LOC646903
|
uncharacterized LOC646903
|
|
chr11_+_69924407
|
2.388
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chrX_+_49687212
|
2.386
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr16_+_56965747
|
2.363
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr10_-_44070008
|
2.362
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr8_+_67341244
|
2.361
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr17_-_74721923
|
2.354
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr17_+_1958354
|
2.354
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr6_+_144164472
|
2.348
|
NM_032860
|
LTV1
|
LTV1 homolog (S. cerevisiae)
|
|
chr12_-_77459198
|
2.344
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr8_+_1950304
|
2.325
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr1_-_6557448
|
2.317
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr3_-_179169330
|
2.314
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr10_+_102747291
|
2.310
|
NM_001163812
NM_001163813
NM_001163814
NM_021830
|
C10orf2
|
chromosome 10 open reading frame 2
|
|
chr6_+_31783530
|
2.296
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr3_-_145878775
|
2.280
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr17_-_43510279
|
2.276
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr5_+_121297655
|
2.272
|
NM_152546
|
SRFBP1
|
serum response factor binding protein 1
|
|
chr9_+_139247508
|
2.269
|
NM_001145639
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr8_-_23261675
|
2.249
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr2_+_88991166
|
2.240
|
NM_144563
|
RPIA
|
ribose 5-phosphate isomerase A
|
|
chr1_+_41445402
|
2.234
|
|
CTPS
|
CTP synthase
|
|
chr12_+_54519836
|
2.232
|
|
LOC400043
|
uncharacterized LOC400043
|
|
chr11_-_798196
|
2.226
|
NM_001191061
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr1_+_41445374
|
2.223
|
|
CTPS
|
CTP synthase
|
|
chr2_-_40006185
|
2.214
|
|
THUMPD2
|
THUMP domain containing 2
|
|
chr6_-_97345676
|
2.213
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr17_+_40985416
|
2.191
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr1_+_763045
|
2.182
|
|
LOC643837
|
uncharacterized LOC643837
|
|
chr12_-_48152180
|
2.182
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr19_-_4670375
|
2.173
|
NM_019107
|
C19orf10
|
chromosome 19 open reading frame 10
|
|
chr1_+_26496382
|
2.171
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr9_+_132388404
|
2.162
|
NM_014064
|
METTL11A
|
methyltransferase like 11A
|
|
chr1_+_41445359
|
2.158
|
|
CTPS
|
CTP synthase
|
|
chr21_+_38071432
|
2.158
|
|
|
|
|
chr5_-_154317653
|
2.153
|
NM_001252156
NM_015465
|
GEMIN5
|
gem (nuclear organelle) associated protein 5
|
|
chr19_-_1237840
|
2.144
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr1_+_19578288
|
2.142
|
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae)
|
|
chr14_+_103995535
|
2.131
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chrX_+_38663072
|
2.128
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr4_-_4291889
|
2.123
|
NM_017816
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr17_+_45000493
|
2.121
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr9_+_132388451
|
2.113
|
|
METTL11A
|
methyltransferase like 11A
|
|
chr21_-_30257611
|
2.109
|
NM_013240
NM_182749
|
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative)
|
|
chr1_+_65613771
|
2.109
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr3_-_81810618
|
2.101
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr8_+_48920969
|
2.100
|
NM_003350
|
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr3_-_81810914
|
2.079
|
NM_000158
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr3_-_179169180
|
2.055
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr7_-_108096693
|
2.051
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr9_-_2844071
|
2.051
|
|
KIAA0020
|
KIAA0020
|
|
chr13_-_20735182
|
2.045
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr6_-_97345760
|
2.045
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr14_-_69444970
|
2.045
|
|
ACTN1
|
actinin, alpha 1
|
|
chr10_+_14920781
|
2.039
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr16_-_12009568
|
2.029
|
|
|
|
|
chr6_+_144164509
|
2.028
|
|
LTV1
|
LTV1 homolog (S. cerevisiae)
|
|
chr17_+_40985425
|
2.016
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr20_+_42544781
|
2.006
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr11_-_68039446
|
2.003
|
NM_022338
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr1_+_762987
|
1.993
|
|
LOC643837
|
uncharacterized LOC643837
|
|
chr3_-_81810802
|
1.993
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr12_-_6677417
|
1.978
|
NM_001033714
NM_006170
|
NOP2
|
NOP2 nucleolar protein homolog (yeast)
|
|
chr3_-_133614421
|
1.973
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chrX_-_152864621
|
1.970
|
NM_001130997
NM_152274
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr14_+_103995473
|
1.964
|
NM_152307
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr19_+_34287662
|
1.949
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr20_-_62711160
|
1.946
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr11_-_797955
|
1.944
|
NM_001191060
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr2_-_224903933
|
1.941
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chrX_-_153151537
|
1.934
|
|
|
|
|
chr7_+_43622638
|
1.926
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr2_+_153191830
|
1.924
|
|
FMNL2
|
formin-like 2
|
|
chr19_+_55591759
|
1.913
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr2_-_158485155
|
1.912
|
|
ACVR1C
|
activin A receptor, type IC
|
|
chr22_+_24577443
|
1.910
|
NM_019601
|
SUSD2
|
sushi domain containing 2
|
|
chr2_-_85108202
|
1.909
|
|
C2orf89
|
chromosome 2 open reading frame 89
|
|
chr8_-_30515665
|
1.906
|
NM_002095
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr3_-_73673978
|
1.904
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr19_+_1026582
|
1.898
|
|
CNN2
|
calponin 2
|
|
chr3_-_81810816
|
1.891
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr3_-_50374749
|
1.891
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr21_-_44299605
|
1.886
|
|
WDR4
|
WD repeat domain 4
|
|
chr11_-_68039327
|
1.882
|
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr17_+_40985434
|
1.880
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr12_-_76424474
|
1.873
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr1_+_101702304
|
1.867
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr8_-_8750739
|
1.865
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr17_+_21279667
|
1.864
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr14_+_24769116
|
1.862
|
|
|
|
|
chr3_-_156272909
|
1.862
|
NM_007107
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr1_+_245318245
|
1.859
|
NM_018012
|
KIF26B
|
kinesin family member 26B
|