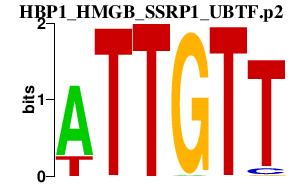
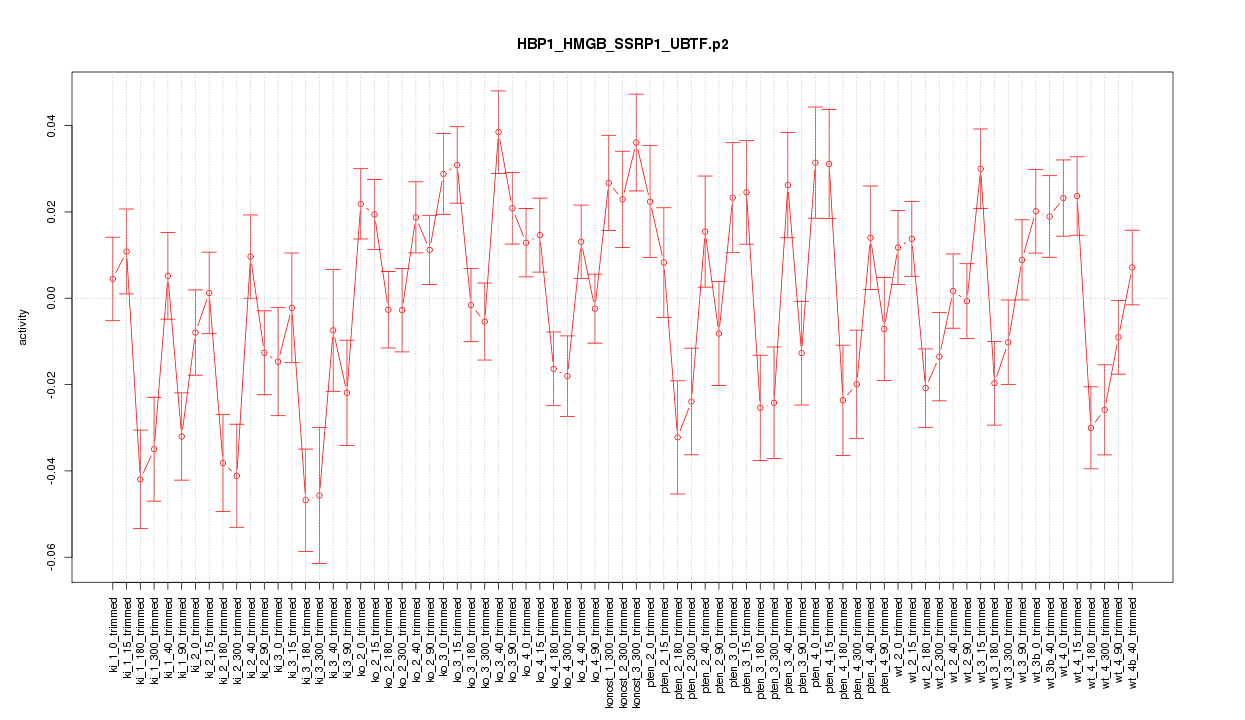
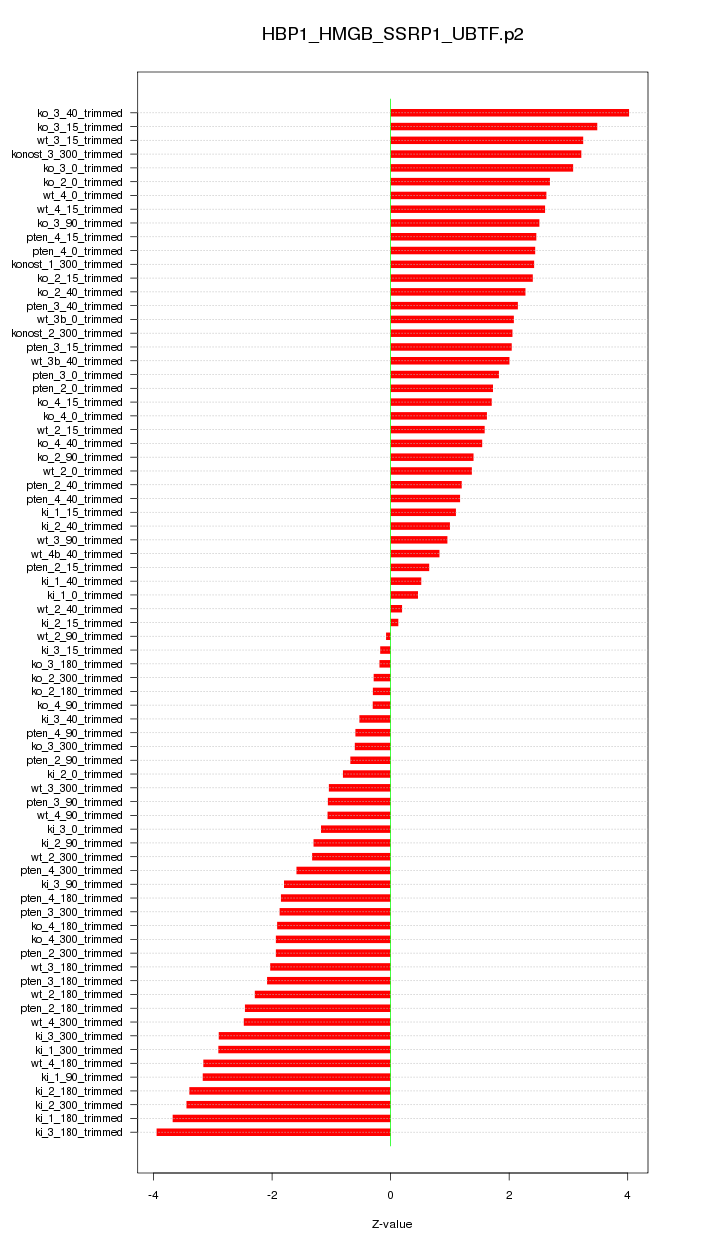
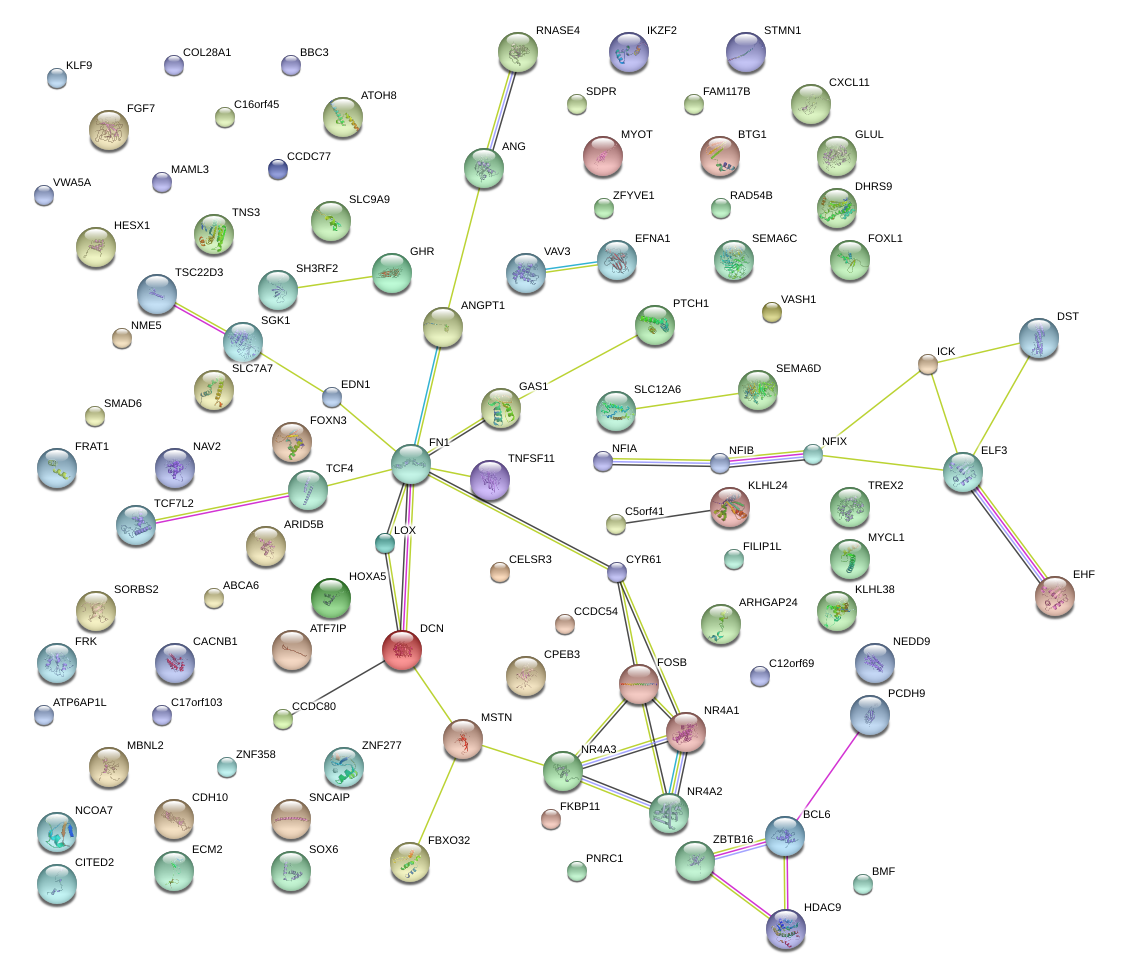
|
chr11_-_16424391
|
46.163
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_-_124553448
|
45.337
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr8_-_124553253
|
38.779
|
|
FBXO32
|
F-box protein 32
|
|
chr19_+_45971249
|
37.966
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr8_-_108510094
|
30.124
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr16_+_86612114
|
27.812
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr6_+_89790464
|
26.986
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr6_+_89790412
|
26.899
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr15_+_48010685
|
25.035
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr18_+_3594056
|
23.366
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr1_-_108231122
|
21.980
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr15_+_66994669
|
20.771
|
NM_005585
|
SMAD6
|
SMAD family member 6
|
|
chr12_-_14967091
|
20.430
|
NM_001013698
|
C12orf69
|
chromosome 12 open reading frame 69
|
|
chr3_-_187454247
|
18.976
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_+_52445185
|
18.717
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr2_-_157189040
|
18.425
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr11_+_34654010
|
17.943
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr6_-_134498973
|
16.308
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_12290528
|
16.014
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr4_+_86699858
|
15.866
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_+_121647773
|
15.278
|
NM_001242935
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr11_+_20049128
|
15.228
|
|
NAV2
|
neuron navigator 2
|
|
chr12_-_91573264
|
15.193
|
NM_133503
|
DCN
|
decorin
|
|
chr2_-_216245573
|
15.180
|
|
FN1
|
fibronectin 1
|
|
chr5_+_81601165
|
14.760
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr10_+_63661012
|
14.593
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr15_-_40401041
|
14.581
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr9_-_95298251
|
14.243
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr2_+_169923531
|
14.091
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr7_-_7575449
|
14.038
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr12_-_122240827
|
13.739
|
|
|
|
|
chr4_-_186877413
|
13.721
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_121814781
|
13.658
|
|
|
|
|
chr5_+_172483285
|
13.500
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr1_-_40367539
|
13.427
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr9_-_73028278
|
13.310
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr6_-_134639195
|
12.961
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_-_24645084
|
12.869
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr8_-_124665165
|
12.707
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr12_-_91576579
|
12.666
|
|
DCN
|
decorin
|
|
chr1_+_86046434
|
12.602
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr15_-_40398573
|
12.533
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr9_-_98268866
|
12.522
|
|
PTCH1
|
patched 1
|
|
chr2_-_216259197
|
12.379
|
|
FN1
|
fibronectin 1
|
|
chr12_+_14538232
|
12.369
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr14_-_89883306
|
12.330
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr17_-_67138013
|
12.319
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr6_-_134496833
|
12.238
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_-_34610928
|
12.209
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr5_+_137203559
|
12.123
|
|
MYOT
|
myotilin
|
|
chr2_-_216246961
|
12.109
|
|
FN1
|
fibronectin 1
|
|
chr1_+_201979689
|
12.033
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr15_+_49715374
|
11.508
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr7_+_27224163
|
11.398
|
|
|
|
|
chr4_-_186732047
|
11.330
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_-_27183225
|
11.246
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chrX_-_106959597
|
11.161
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_123986103
|
11.152
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr13_-_67804432
|
11.074
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr3_-_99594915
|
11.055
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr19_+_7580987
|
11.018
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr6_-_139695739
|
10.921
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr6_-_139695784
|
10.847
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr10_+_99079017
|
10.837
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr9_-_14313509
|
10.711
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_155099935
|
10.617
|
|
EFNA1
|
ephrin-A1
|
|
chr16_+_15528324
|
10.604
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr6_-_139695349
|
10.517
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr1_-_26233367
|
10.408
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr12_-_92539614
|
10.332
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr5_-_121414011
|
10.126
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr17_-_21156577
|
10.077
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr4_-_186577858
|
9.976
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_126240369
|
9.897
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr3_-_112359547
|
9.876
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_201476283
|
9.859
|
|
|
|
|
chr3_-_57234279
|
9.857
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr3_-_112359443
|
9.849
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_183353355
|
9.829
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr9_-_89562103
|
9.778
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr9_-_14314036
|
9.723
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_112359565
|
9.693
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr9_-_14313899
|
9.687
|
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_113931228
|
9.566
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr7_-_142985270
|
9.482
|
|
|
|
|
chr13_+_97794050
|
9.458
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr4_-_141074122
|
9.435
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr5_+_137203544
|
9.428
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr3_+_107096187
|
9.362
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr11_+_19372270
|
9.292
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr6_-_116381765
|
9.269
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr1_+_201979747
|
9.220
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr5_-_137475076
|
9.163
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr3_-_143567163
|
9.132
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr14_-_73493744
|
9.089
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr6_-_134497069
|
8.993
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_-_186732208
|
8.936
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_20044362
|
8.925
|
|
NAV2
|
neuron navigator 2
|
|
chr6_-_11232883
|
8.766
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_-_190927321
|
8.765
|
NM_005259
|
MSTN
|
myostatin
|
|
chr14_-_23288912
|
8.758
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr6_-_11232901
|
8.670
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_+_42565963
|
8.633
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr5_-_121413906
|
8.583
|
|
LOX
|
lysyl oxidase
|
|
chr12_+_498513
|
8.580
|
NM_001130147
NM_001130148
|
CCDC77
|
coiled-coil domain containing 77
|
|
chr7_+_18535351
|
8.569
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr11_+_34642651
|
8.547
|
|
EHF
|
ets homologous factor
|
|
chr10_-_94003002
|
8.543
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr6_-_52926588
|
8.503
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr7_-_47621635
|
8.490
|
|
TNS3
|
tensin 3
|
|
chr3_-_48700317
|
8.430
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr1_-_182360181
|
8.388
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_-_121413967
|
8.352
|
|
LOX
|
lysyl oxidase
|
|
chr9_+_127420690
|
8.351
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr1_-_16161279
|
8.233
|
|
|
|
|
chr6_-_11232900
|
8.221
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr14_+_21156922
|
8.202
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr2_-_214015053
|
8.164
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_+_85980600
|
8.153
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr7_+_111846642
|
8.123
|
NM_021994
|
ZNF277
|
zinc finger protein 277
|
|
chr1_-_182360400
|
8.104
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr6_-_56716412
|
8.072
|
|
DST
|
dystonin
|
|
chr7_+_111846736
|
8.039
|
|
ZNF277
|
zinc finger protein 277
|
|
chr13_+_43136871
|
7.896
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr18_-_29993195
|
7.883
|
|
|
|
|
chr4_-_76957139
|
7.881
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr7_-_47621741
|
7.845
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr19_-_47734215
|
7.819
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr6_-_56716713
|
7.763
|
|
DST
|
dystonin
|
|
chr14_+_77228940
|
7.733
|
|
VASH1
|
vasohibin 1
|
|
chr1_-_151118932
|
7.707
|
NM_001178061
NM_001178062
NM_030913
|
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C
|
|
chr2_+_203499667
|
7.613
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr2_-_56412904
|
7.610
|
|
LOC100129434
|
uncharacterized LOC100129434
|
|
chr2_-_192711651
|
7.566
|
|
SDPR
|
serum deprivation response
|
|
chr5_+_145317297
|
7.564
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr9_-_14086901
|
7.502
|
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_95449131
|
7.462
|
NM_001205263
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr18_-_52988916
|
7.445
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr12_+_7169192
|
7.418
|
|
|
|
|
chr4_-_141075232
|
7.404
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr2_-_26205437
|
7.392
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr15_-_41166336
|
7.388
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr2_-_122159140
|
7.364
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr17_-_46035109
|
7.363
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr2_-_26205327
|
7.347
|
|
KIF3C
|
kinesin family member 3C
|
|
chr16_-_30934437
|
7.344
|
|
FBXL19-AS1
|
FBXL19 antisense RNA 1 (non-protein coding)
|
|
chr1_+_51435911
|
7.199
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr11_+_34642587
|
7.184
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr2_-_211035377
|
7.136
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr11_-_18258315
|
7.135
|
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr1_-_111970354
|
7.130
|
NM_002557
|
OVGP1
|
oviductal glycoprotein 1, 120kDa
|
|
chr3_-_15563160
|
7.103
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr12_+_9142136
|
7.088
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr12_-_62586547
|
7.062
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr22_-_50970542
|
7.031
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr6_+_43027628
|
7.027
|
NM_201523
|
KLC4
|
kinesin light chain 4
|
|
chr6_-_52926486
|
7.017
|
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr8_+_72756218
|
7.003
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr6_-_152957985
|
6.950
|
NM_033071
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr4_-_186578122
|
6.939
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr19_+_36132646
|
6.922
|
NM_014209
|
ETV2
|
ets variant 2
|
|
chr5_+_121465214
|
6.912
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr3_+_189507448
|
6.886
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr4_+_39063851
|
6.845
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr5_+_78532338
|
6.836
|
|
|
|
|
chr3_+_101546833
|
6.820
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr12_-_10562142
|
6.801
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 readthrough
killer cell lectin-like receptor subfamily C, member 4
|
|
chr2_-_165424993
|
6.795
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr11_-_85430117
|
6.743
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_+_109625347
|
6.736
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr20_-_61002588
|
6.733
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr10_-_62332666
|
6.731
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_-_18480203
|
6.729
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr2_-_192711923
|
6.724
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr2_-_175869910
|
6.722
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr1_-_116383323
|
6.721
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr18_+_55888750
|
6.707
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_+_41323181
|
6.698
|
NM_005899
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr18_+_3451589
|
6.686
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr14_-_92413738
|
6.673
|
|
FBLN5
|
fibulin 5
|
|
chr2_+_33359607
|
6.671
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr19_-_40324747
|
6.662
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr13_-_33780142
|
6.643
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_164528914
|
6.637
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr7_-_107643330
|
6.604
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr3_+_69985750
|
6.601
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_23826062
|
6.599
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_182357757
|
6.548
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr14_-_55878537
|
6.446
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr18_+_7946885
|
6.445
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr4_-_52883785
|
6.427
|
NM_001024611
|
LRRC66
|
leucine rich repeat containing 66
|
|
chr11_-_16497917
|
6.427
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_-_214016332
|
6.346
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr3_+_160394946
|
6.310
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr14_-_24020766
|
6.254
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr18_+_55888838
|
6.250
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_163099877
|
6.241
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr1_-_53018721
|
6.209
|
NM_001009881
NM_015269
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr13_-_48987017
|
6.207
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr7_-_27187367
|
6.189
|
NM_024014
|
HOXA6
|
homeobox A6
|