|
chr3_-_87040268
|
2.296
|
|
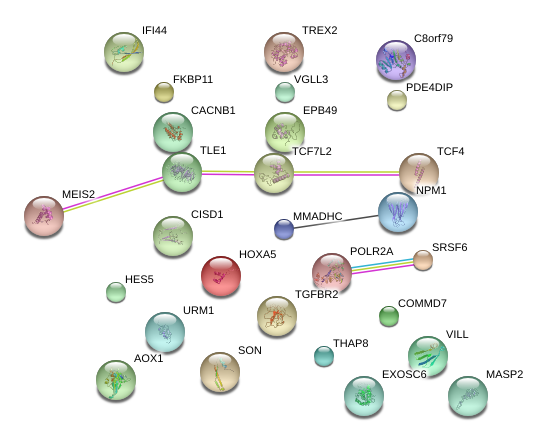
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr3_-_87040246
|
2.219
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr17_-_70588942
|
0.638
|
|
LOC100499467
|
uncharacterized LOC100499467
|
|
chr1_+_79115476
|
0.423
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr2_+_201450730
|
0.416
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr7_-_27183225
|
0.296
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr2_+_201450579
|
0.283
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr3_+_38032260
|
0.246
|
|
VILL
|
villin-like
|
|
chr8_+_12869772
|
0.170
|
NM_001099677
|
KIAA1456
|
KIAA1456
|
|
chr19_-_36545298
|
0.129
|
|
THAP8
|
THAP domain containing 8
|
|
chr1_-_144994908
|
0.118
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_11107295
|
0.098
|
NM_006610
NM_139208
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr9_-_84304378
|
0.095
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr19_-_36545119
|
0.095
|
|
THAP8
|
THAP domain containing 8
|
|
chr2_-_150444285
|
0.088
|
NM_015702
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr16_-_70285794
|
0.080
|
NM_058219
|
EXOSC6
|
exosome component 6
|
|
chr1_-_2461683
|
0.075
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr18_-_53303129
|
0.073
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr3_+_30648372
|
0.053
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr2_-_150444236
|
0.052
|
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria
|
|
chr8_+_21916685
|
0.049
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr20_-_31331196
|
0.045
|
|
COMMD7
|
COMM domain containing 7
|
|
chr1_-_144994839
|
0.041
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr9_+_131133597
|
0.040
|
NM_001135947
NM_030914
|
URM1
|
ubiquitin related modifier 1
|
|
chr3_+_30647901
|
0.033
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr20_+_42086533
|
0.031
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr15_-_37393364
|
0.030
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr17_+_7387688
|
0.023
|
NM_000937
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa
|
|
chr1_-_144994943
|
0.022
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr5_+_170814707
|
0.013
|
NM_001037738
NM_002520
NM_199185
|
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin)
|
|
chr10_+_60028885
|
0.008
|
NM_018464
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
chr21_+_34922923
|
0.003
|
|
SON
|
SON DNA binding protein
|
|
chr1_-_11107150
|
0.002
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|