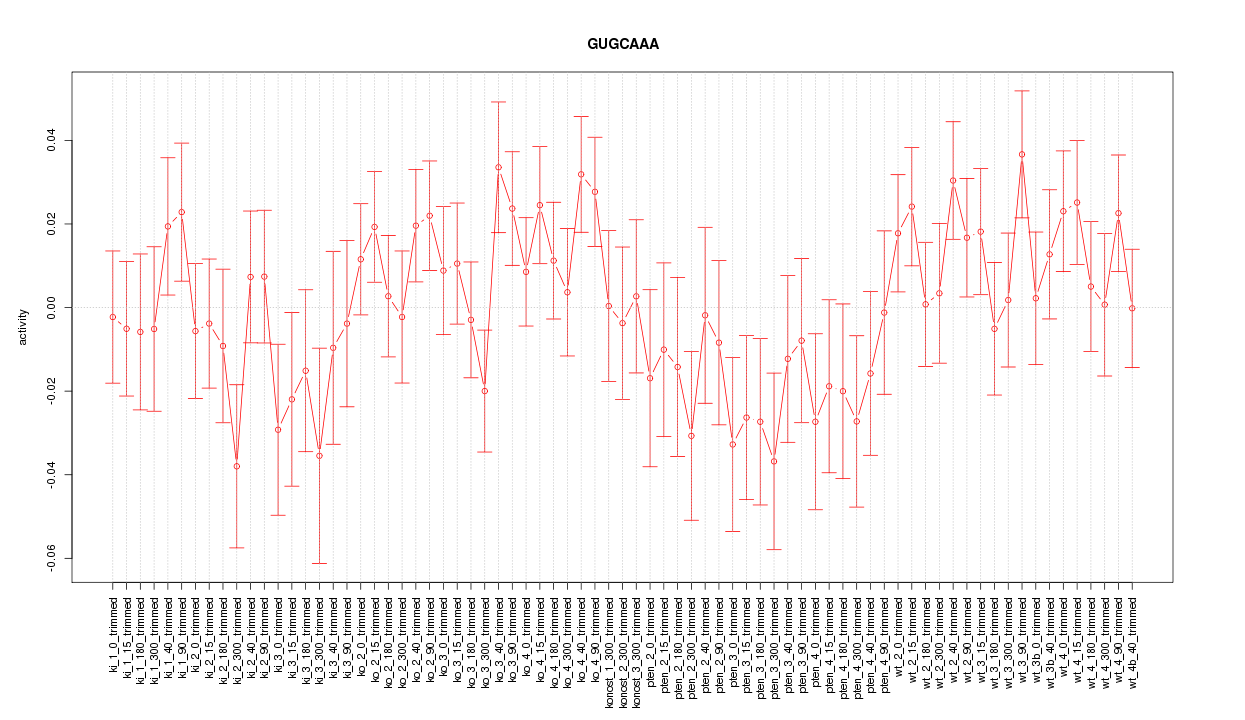
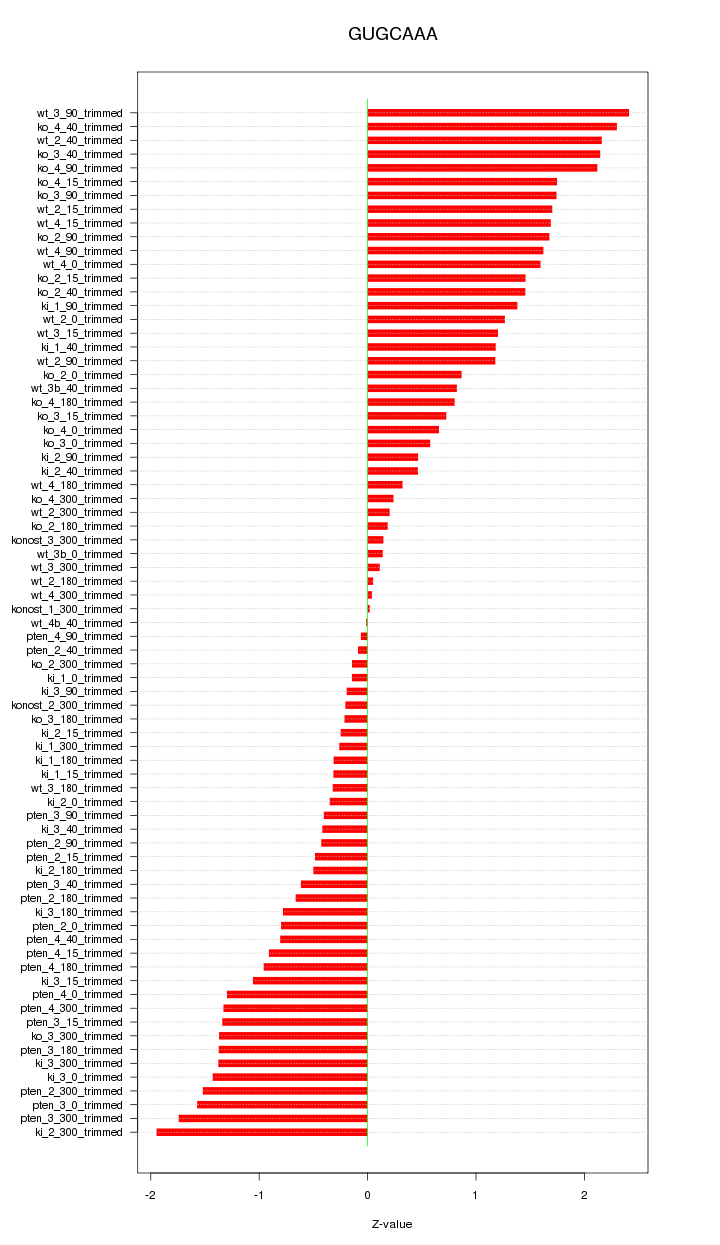
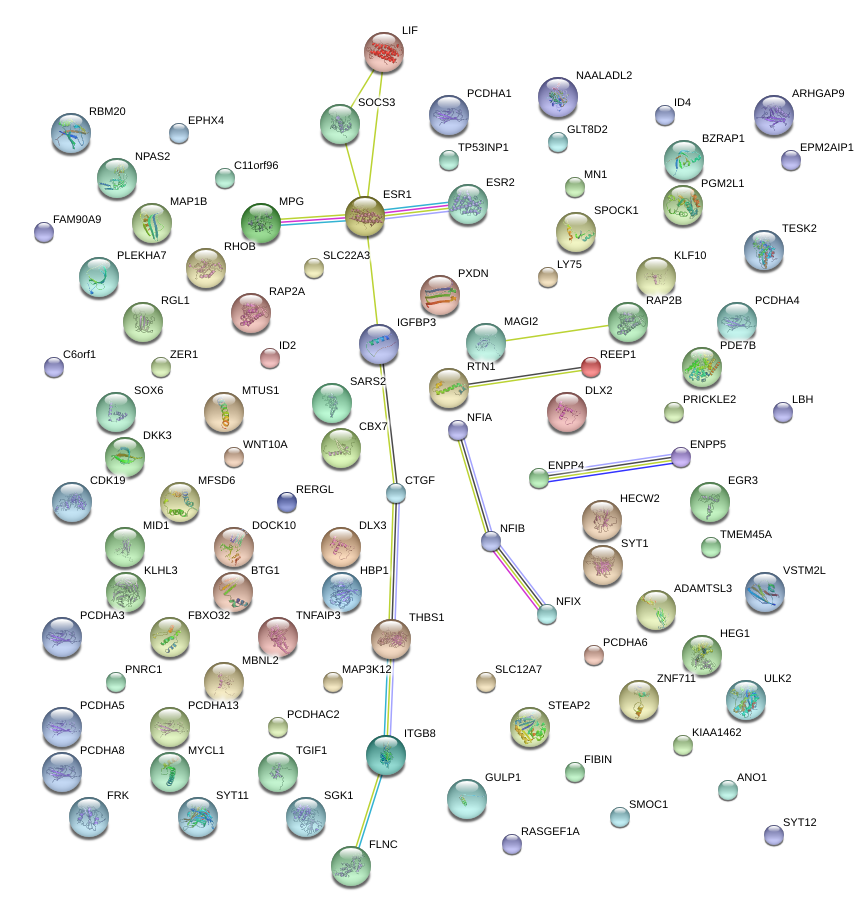
|
chr2_+_30454396
|
16.906
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr11_+_69924407
|
10.781
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr11_+_43963809
|
9.258
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr1_+_92495484
|
8.830
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr8_-_22550057
|
8.445
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
8.329
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
8.088
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr3_-_64211111
|
7.964
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr8_-_124544376
|
7.589
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr2_-_1748285
|
7.290
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr8_-_124553448
|
6.984
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr5_+_140254930
|
6.570
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr7_+_128470462
|
6.467
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr6_-_132272311
|
6.284
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr5_-_136834887
|
6.081
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr17_-_48072566
|
5.672
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr2_-_86564632
|
5.354
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr11_-_12030128
|
5.232
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr2_+_219745243
|
5.097
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr6_+_160769365
|
4.849
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr2_-_197457250
|
4.755
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr22_-_30642683
|
4.570
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr3_-_124774735
|
4.433
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr10_-_30348433
|
4.417
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr7_-_45960738
|
4.383
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr22_-_39548448
|
4.348
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr14_+_70346113
|
4.323
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr17_-_19770988
|
4.263
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr1_-_40367539
|
4.262
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr15_+_39873276
|
3.925
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr17_-_76356153
|
3.855
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr5_+_140165875
|
3.752
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr20_+_36531498
|
3.732
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chrX_+_84498993
|
3.717
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr6_+_46097700
|
3.629
|
NM_014936
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr6_+_136172802
|
3.607
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_225907312
|
3.584
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr2_+_20646831
|
3.458
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr6_-_134497069
|
3.431
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_+_79439432
|
3.420
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr5_+_140247767
|
3.383
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr2_-_160761266
|
3.338
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr6_-_134498973
|
3.324
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_45956795
|
3.324
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr6_+_89790412
|
3.207
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr6_-_134496006
|
3.185
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_189156395
|
3.182
|
NM_001252668
NM_001252669
NM_016315
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr12_+_79257772
|
3.167
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr6_-_46138584
|
3.134
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr5_-_1112106
|
3.111
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr12_+_79258448
|
3.094
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_-_137071703
|
3.090
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr5_+_140235594
|
3.050
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr6_-_134639195
|
3.017
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_174577069
|
3.010
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr8_-_17658385
|
2.970
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_+_19837599
|
2.887
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr11_-_74109417
|
2.874
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr9_-_131534186
|
2.859
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr5_+_140207562
|
2.847
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr2_+_191273012
|
2.836
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr7_-_79082870
|
2.833
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_+_27015627
|
2.827
|
NM_203371
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr12_-_53893195
|
2.811
|
NM_001193511
NM_006301
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chr12_-_92539614
|
2.806
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr18_+_3453771
|
2.785
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_-_116381765
|
2.784
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr8_-_95961543
|
2.778
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr8_-_103667882
|
2.723
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr18_+_3447607
|
2.719
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr5_+_140345746
|
2.717
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr18_+_3412071
|
2.696
|
NM_174886
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr14_-_60337350
|
2.694
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr9_-_14398981
|
2.677
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_155829259
|
2.653
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr2_+_8822112
|
2.634
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr15_+_84322809
|
2.632
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr3_+_152880024
|
2.618
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr7_+_20370724
|
2.578
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr13_+_97874540
|
2.554
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr17_-_56406120
|
2.534
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr18_+_3449972
|
2.502
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr9_-_14314036
|
2.494
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr10_-_43762366
|
2.491
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr1_+_183605207
|
2.485
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_-_111136407
|
2.476
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr5_+_140306301
|
2.460
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr10_+_112404104
|
2.454
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr11_-_16424391
|
2.424
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_17035948
|
2.415
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr2_+_101436491
|
2.349
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr8_-_103666018
|
2.348
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr5_+_140180782
|
2.322
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr22_-_28197469
|
2.301
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr11_-_12030882
|
2.278
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr3_+_100211408
|
2.273
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr7_+_106809405
|
2.221
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr18_+_3451589
|
2.175
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_+_138188352
|
2.175
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr7_+_89840999
|
2.171
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr5_+_71403040
|
2.164
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_+_140186658
|
2.158
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr6_+_152128685
|
2.151
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr11_-_12030622
|
2.116
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr5_+_140201221
|
2.019
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chrX_-_10645726
|
1.920
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_-_16430425
|
1.911
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr10_+_97889467
|
1.876
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr5_+_140220811
|
1.871
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr8_-_110657025
|
1.871
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_-_86565205
|
1.867
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr8_-_110704019
|
1.864
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr7_-_112579726
|
1.827
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr10_+_89623091
|
1.823
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr6_+_121756744
|
1.811
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr8_-_110661007
|
1.795
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_-_16497917
|
1.784
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_+_151483861
|
1.784
|
NM_020770
|
CGN
|
cingulin
|
|
chr3_-_18466759
|
1.773
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr3_+_50712671
|
1.765
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr6_+_21593963
|
1.729
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr5_+_140213968
|
1.729
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr8_-_110657745
|
1.717
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_-_22697439
|
1.713
|
NM_014802
|
KIAA0528
|
KIAA0528
|
|
chr11_+_121322848
|
1.674
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_+_207627614
|
1.667
|
NM_001006658
NM_001877
|
CR2
|
complement component (3d/Epstein Barr virus) receptor 2
|
|
chrX_-_10588458
|
1.667
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_+_77089248
|
1.666
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr8_-_110693233
|
1.664
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr4_-_122854192
|
1.615
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr16_-_14788429
|
1.587
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr20_-_56286540
|
1.583
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr20_-_56285576
|
1.580
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr20_+_8112762
|
1.576
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr17_-_61777478
|
1.566
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr3_+_184279539
|
1.547
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr4_+_87856152
|
1.536
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr10_+_23728197
|
1.504
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr10_-_61666236
|
1.482
|
NM_005436
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr1_+_6673744
|
1.465
|
NM_153812
|
PHF13
|
PHD finger protein 13
|
|
chr8_-_110655418
|
1.461
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_+_118401755
|
1.448
|
NM_001144034
NM_001144035
NM_001144036
NM_001144037
NM_001144038
NM_032780
|
TMEM25
|
transmembrane protein 25
|
|
chr15_+_48010685
|
1.446
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr14_+_59655351
|
1.432
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr3_-_18480203
|
1.427
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr2_+_198669316
|
1.426
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr16_+_4382224
|
1.416
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr3_+_43732358
|
1.408
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr5_-_158637060
|
1.407
|
NM_001199380
|
RNF145
|
ring finger protein 145
|
|
chr1_-_214724974
|
1.397
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr12_-_64616032
|
1.385
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr11_+_8060179
|
1.383
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr12_+_12938540
|
1.380
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr7_-_98741681
|
1.371
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr2_-_182545212
|
1.359
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr4_+_87928083
|
1.358
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr5_-_100238870
|
1.353
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chrX_+_44732402
|
1.351
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chrX_-_10851808
|
1.351
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_-_111021026
|
1.343
|
NM_139283
|
PPTC7
|
PTC7 protein phosphatase homolog (S. cerevisiae)
|
|
chr5_-_158636489
|
1.340
|
NM_001199383
|
RNF145
|
ring finger protein 145
|
|
chr16_+_50775960
|
1.334
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr11_-_8190535
|
1.322
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr7_-_14025826
|
1.319
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr17_+_45331154
|
1.315
|
NM_000212
|
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61)
|
|
chr1_-_160232286
|
1.314
|
NM_015726
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr9_-_23821477
|
1.304
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_+_17653412
|
1.299
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr9_-_23826062
|
1.293
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr16_-_11350038
|
1.283
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr1_+_42846427
|
1.282
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr3_-_184870727
|
1.281
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr16_+_50776012
|
1.269
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr14_+_65007163
|
1.262
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr6_-_16761600
|
1.257
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr8_-_12973752
|
1.242
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_+_102153858
|
1.242
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr17_-_73851377
|
1.240
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr20_-_3388219
|
1.230
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr8_-_12990731
|
1.226
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_+_152011630
|
1.221
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr3_-_87040246
|
1.220
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr14_+_52781004
|
1.214
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr10_+_63661012
|
1.211
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr9_-_23821842
|
1.202
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr20_-_5591487
|
1.193
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chrX_-_10544852
|
1.192
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr5_+_140261792
|
1.190
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr11_+_111848008
|
1.185
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr7_-_75368276
|
1.169
|
NM_001243198
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chr2_-_208890207
|
1.148
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr7_-_105319608
|
1.143
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr15_+_98503907
|
1.138
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr5_-_158635080
|
1.137
|
NM_001199381
|
RNF145
|
ring finger protein 145
|
|
chr19_+_11200037
|
1.136
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr12_-_120315074
|
1.132
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr2_-_68694389
|
1.132
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chrX_+_17393537
|
1.110
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr11_+_111807857
|
1.104
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr8_-_143695812
|
1.100
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|