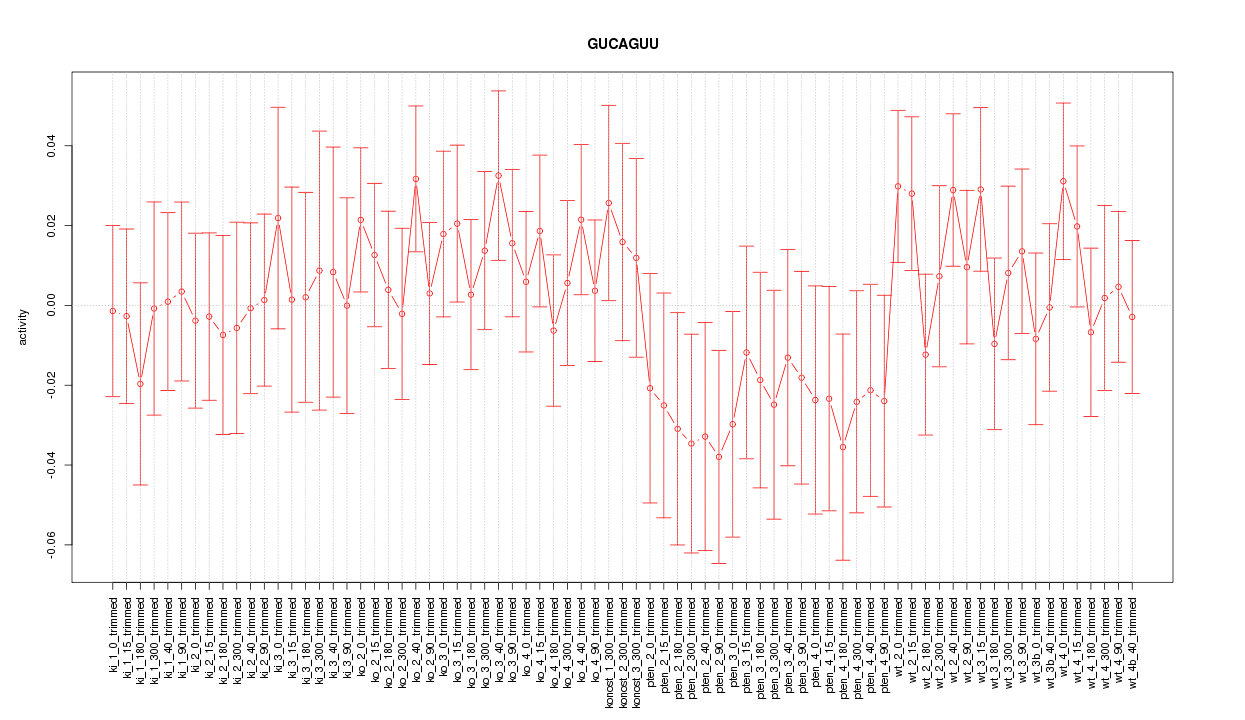
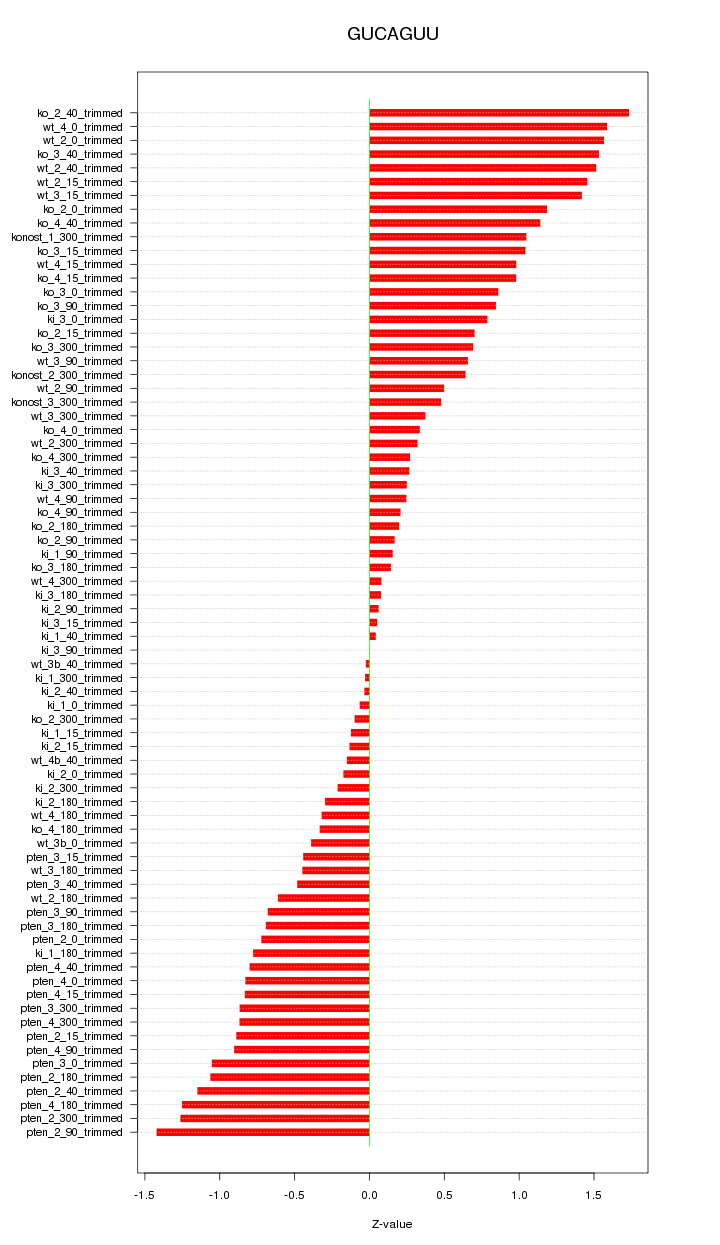
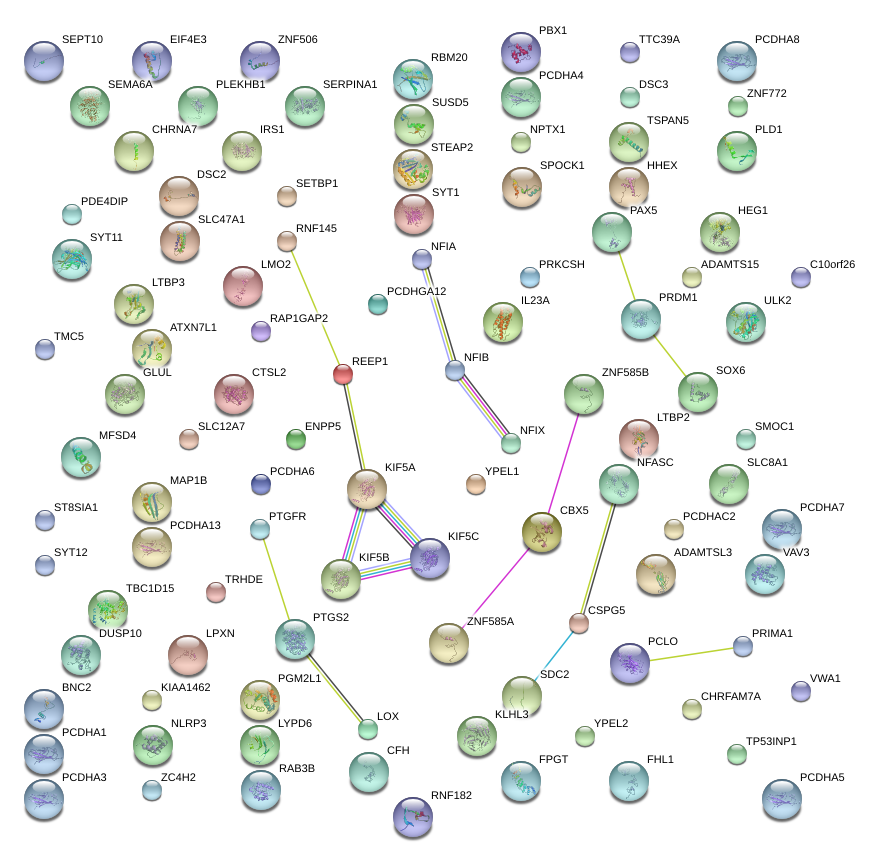
|
chr9_-_99801365
|
6.859
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr9_-_99801924
|
6.663
|
NM_001201575
|
CTSL2
|
cathepsin L2
|
|
chr17_-_19770988
|
4.546
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr4_-_99579540
|
4.361
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr5_+_140254930
|
4.250
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr17_+_19437140
|
4.013
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr19_-_37701385
|
3.912
|
NM_152279
|
ZNF585B
|
zinc finger protein 585B
|
|
chr19_-_37663567
|
3.805
|
NM_152655
|
ZNF585A
|
zinc finger protein 585A
|
|
chr5_-_1112106
|
3.650
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chrX_-_64196268
|
3.645
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr7_+_89840999
|
3.187
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr2_-_86564632
|
2.757
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr19_-_19932512
|
2.627
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr6_+_13925097
|
2.598
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chrX_-_64254592
|
2.459
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr19_-_37663332
|
2.355
|
NM_199126
|
ZNF585A
|
zinc finger protein 585A
|
|
chr5_+_140247767
|
2.229
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr6_+_13925423
|
2.158
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr5_+_140777603
|
2.145
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr2_+_150187028
|
2.039
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr11_+_66790815
|
2.034
|
NM_177963
|
SYT12
|
synaptotagmin XII
|
|
chr10_+_94449678
|
1.976
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr5_+_140165875
|
1.924
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr2_+_150186498
|
1.911
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr6_+_106546736
|
1.846
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr7_-_82792127
|
1.808
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr5_+_140235594
|
1.797
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr6_+_106534188
|
1.771
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_+_140345746
|
1.769
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr10_+_112404104
|
1.764
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr14_-_94254765
|
1.722
|
NM_178013
|
PRIMA1
|
proline rich membrane anchor 1
|
|
chr11_+_130318457
|
1.698
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr6_-_46138584
|
1.687
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr14_-_75078669
|
1.652
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr11_-_33913835
|
1.606
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr5_+_140306301
|
1.598
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr5_-_115910406
|
1.583
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr1_-_108507474
|
1.577
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr5_+_140207562
|
1.536
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr5_+_140180782
|
1.520
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr11_+_66790189
|
1.516
|
NM_001177880
|
SYT12
|
synaptotagmin XII
|
|
chr19_-_57988891
|
1.509
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr10_-_30348433
|
1.493
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr11_-_33891361
|
1.472
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr5_+_71403040
|
1.457
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_58343308
|
1.453
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr1_-_52456292
|
1.417
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr5_+_140186658
|
1.413
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr11_-_58345593
|
1.409
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr2_-_40657419
|
1.400
|
NM_001112800
NM_001112801
NM_001252624
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr5_+_140201221
|
1.399
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr11_-_33891485
|
1.398
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr5_+_140220811
|
1.360
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr11_+_73358593
|
1.358
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chrX_+_135278912
|
1.356
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_135229536
|
1.353
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_1535446
|
1.338
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr3_-_33260706
|
1.338
|
NM_015551
|
SUSD5
|
sushi domain containing 5
|
|
chr5_-_136834887
|
1.336
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr12_-_22487566
|
1.330
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr1_-_108231122
|
1.317
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_-_186649421
|
1.315
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr14_+_70346113
|
1.307
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr5_+_140213968
|
1.296
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr6_+_13924676
|
1.291
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr2_-_86565205
|
1.283
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr15_+_84322809
|
1.218
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr1_+_247581350
|
1.177
|
NM_001127462
NM_001243133
NM_004895
NM_183395
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chrX_+_135230736
|
1.163
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_74109417
|
1.158
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr1_-_51810693
|
1.130
|
NM_001144832
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr14_-_94856926
|
1.106
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr8_+_97505876
|
1.106
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr1_-_221915458
|
1.096
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_+_72666462
|
1.087
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr7_-_105319608
|
1.081
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr17_-_78450247
|
1.053
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr3_-_171528227
|
1.034
|
NM_001130081
NM_002662
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific
|
|
chr3_-_124774735
|
1.025
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr14_-_94855120
|
1.021
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_221910801
|
1.008
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr2_-_227663454
|
1.002
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_16870719
|
0.992
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr12_+_79258448
|
0.984
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr8_-_95961543
|
0.981
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr12_+_72233486
|
0.980
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr1_+_1370902
|
0.977
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr1_+_247579457
|
0.973
|
NM_001079821
NM_001127461
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr5_-_158636489
|
0.959
|
NM_001199383
|
RNF145
|
ring finger protein 145
|
|
chr1_+_78956727
|
0.957
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr2_-_110371669
|
0.957
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr12_+_79257772
|
0.954
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79439432
|
0.947
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr15_+_32322690
|
0.944
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr1_-_145075636
|
0.943
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_51787881
|
0.941
|
NM_001080494
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr1_+_74701070
|
0.941
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chrX_+_135251454
|
0.935
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_149632772
|
0.934
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr16_+_19422056
|
0.930
|
NM_001105248
NM_001105249
|
TMC5
|
transmembrane channel-like 5
|
|
chr5_-_121414011
|
0.928
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr5_-_158637060
|
0.926
|
NM_001199380
|
RNF145
|
ring finger protein 145
|
|
chr12_+_56732658
|
0.924
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr5_-_121412805
|
0.920
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr5_-_137071703
|
0.918
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr1_+_204797774
|
0.914
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chrX_+_135251795
|
0.914
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_16430425
|
0.913
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr17_+_2699731
|
0.904
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr16_+_19467771
|
0.883
|
NM_024780
|
TMC5
|
transmembrane channel-like 5
|
|
chr5_+_140767402
|
0.880
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr5_+_140810126
|
0.871
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_-_182361311
|
0.864
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr12_-_54673901
|
0.864
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr3_-_47620186
|
0.861
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr1_-_182360400
|
0.857
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr9_-_14398981
|
0.850
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr17_+_57408907
|
0.846
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr1_+_164528586
|
0.840
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr18_+_42260774
|
0.831
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr10_+_104503724
|
0.822
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr1_+_205538111
|
0.817
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr3_-_71774407
|
0.816
|
NM_001134651
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr1_+_204913353
|
0.812
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr18_-_28622624
|
0.810
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr15_-_40398286
|
0.798
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr5_+_140261792
|
0.797
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr17_+_70117154
|
0.795
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr9_-_14314036
|
0.782
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr7_-_112726143
|
0.768
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr8_-_67525426
|
0.760
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr2_-_26205437
|
0.758
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr11_-_16497917
|
0.758
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_+_119421865
|
0.754
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr6_-_36355342
|
0.753
|
NM_001207038
NM_001207035
NM_001207036
NM_001207037
NM_001207039
NM_001207040
NM_001207041
NM_016135
|
ETV7
|
ets variant 7
|
|
chr16_+_8768403
|
0.740
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr4_-_90759446
|
0.737
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr1_-_144997110
|
0.718
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr22_-_38577692
|
0.716
|
NM_001004426
NM_003560
NM_001199562
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr4_-_90758126
|
0.715
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr15_+_99192696
|
0.714
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr17_+_40439564
|
0.714
|
NM_003152
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr12_+_133707212
|
0.711
|
NM_015394
|
ZNF10
|
zinc finger protein 10
|
|
chr1_-_21059132
|
0.711
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr3_-_47621729
|
0.702
|
NM_001206942
NM_001206945
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chrX_+_12993224
|
0.699
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr15_-_40398573
|
0.698
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr11_+_73357222
|
0.686
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr10_+_99079017
|
0.686
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr10_+_82168206
|
0.682
|
NM_001243780
NM_001243781
NM_032333
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr3_-_57199402
|
0.671
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr2_-_178483677
|
0.670
|
NM_152275
|
TTC30A
|
tetratricopeptide repeat domain 30A
|
|
chr18_+_51885170
|
0.670
|
NM_173529
|
C18orf54
|
chromosome 18 open reading frame 54
|
|
chr5_-_132948222
|
0.669
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr5_+_140174443
|
0.665
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr20_-_9819406
|
0.654
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr5_-_158634833
|
0.649
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr6_-_79787939
|
0.648
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr1_-_214724974
|
0.634
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr15_-_40401041
|
0.633
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr3_-_71803923
|
0.624
|
NM_173359
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr5_-_158635080
|
0.623
|
NM_001199381
|
RNF145
|
ring finger protein 145
|
|
chr4_-_25161908
|
0.622
|
NM_016955
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase
|
|
chr5_-_9546157
|
0.614
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr16_+_8814567
|
0.612
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_-_18871163
|
0.610
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_-_145277879
|
0.598
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_+_8806825
|
0.598
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr22_-_39548448
|
0.595
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr1_+_42846427
|
0.574
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr22_-_39240016
|
0.572
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chrX_+_57618268
|
0.561
|
NM_007157
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr16_+_30960377
|
0.553
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr13_+_44453968
|
0.552
|
NM_153218
|
LACC1
|
laccase (multicopper oxidoreductase) domain containing 1
|
|
chr17_-_8066254
|
0.535
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr9_-_27529434
|
0.531
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr4_-_175205401
|
0.530
|
NM_012180
|
FBXO8
|
F-box protein 8
|
|
chr14_+_32798478
|
0.528
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr5_+_140718353
|
0.522
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr14_-_80677839
|
0.516
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr12_-_21654484
|
0.516
|
NM_002907
NM_032941
|
RECQL
|
RecQ protein-like (DNA helicase Q1-like)
|
|
chr11_+_14665268
|
0.515
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr9_-_123639566
|
0.513
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr7_-_130080823
|
0.513
|
NM_018718
|
CEP41
|
centrosomal protein 41kDa
|
|
chr7_-_14025826
|
0.511
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_-_39693110
|
0.509
|
NM_145027
|
KIF6
|
kinesin family member 6
|
|
chr14_+_38677186
|
0.503
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr10_-_61122219
|
0.496
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr3_-_71803513
|
0.495
|
NM_001134649
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr10_-_123353771
|
0.489
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_-_48462561
|
0.488
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr7_+_29846112
|
0.487
|
NM_001080529
|
WIPF3
|
WAS/WASL interacting protein family, member 3
|
|
chrX_+_53449804
|
0.486
|
NM_144968
NM_001031745
|
RIBC1
|
RIB43A domain with coiled-coils 1
|
|
chr13_-_110438896
|
0.482
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr2_+_203499819
|
0.482
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr10_+_97889467
|
0.482
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr3_-_184870727
|
0.481
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr5_+_140729723
|
0.477
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr13_-_77460432
|
0.476
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr1_+_118148506
|
0.467
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|