|
chr11_-_119252364
|
5.289
|
NM_001243759
NM_004205
|
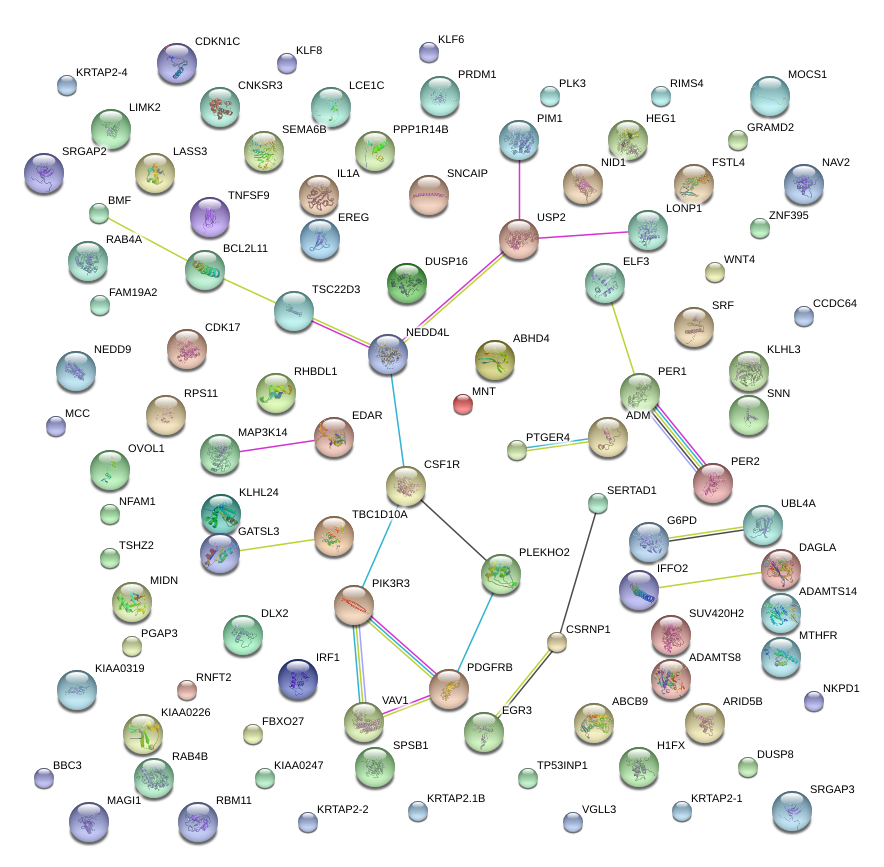
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_-_119234859
|
5.105
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_19282773
|
3.893
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr2_-_239197091
|
3.717
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr15_-_40398573
|
3.498
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr17_-_2304217
|
3.437
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr8_-_22550057
|
3.434
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
3.415
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
3.411
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr15_-_40398286
|
3.377
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr15_-_40401041
|
3.346
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr17_-_8055684
|
3.242
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr1_+_201979689
|
3.199
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr19_+_6772705
|
3.177
|
NM_005428
|
VAV1
|
vav 1 guanine nucleotide exchange factor
|
|
chr5_+_40680021
|
3.168
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr1_+_9352931
|
2.983
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr18_+_55714457
|
2.925
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55888750
|
2.915
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_22469414
|
2.910
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr18_+_55816546
|
2.847
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55862577
|
2.837
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55711388
|
2.837
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_236228476
|
2.822
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr22_-_30685485
|
2.741
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr17_-_39203423
|
2.670
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr11_-_1593149
|
2.627
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr3_-_124774735
|
2.588
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr11_-_2906960
|
2.578
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr11_+_61447816
|
2.499
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr19_-_45663407
|
2.451
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr10_+_63661012
|
2.421
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr19_-_47735858
|
2.409
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr3_-_87040246
|
2.389
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr5_-_137071703
|
2.357
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr5_-_132948222
|
2.231
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr15_-_101084871
|
2.189
|
NM_178842
|
CERS3
|
ceramide synthase 3
|
|
chr19_+_1248548
|
2.179
|
NM_177401
|
MIDN
|
midnolin
|
|
chr2_-_109605668
|
2.157
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr2_-_113542970
|
2.154
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr20_+_51588945
|
2.148
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_-_46598250
|
2.111
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr3_-_39195101
|
2.107
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr12_-_12715244
|
2.099
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr21_+_15588465
|
2.081
|
NM_144770
|
RBM11
|
RNA binding motif protein 11
|
|
chr16_+_726074
|
2.069
|
NM_003961
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr5_-_131826426
|
2.040
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr11_+_19734821
|
2.037
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr19_-_40931880
|
2.004
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr6_-_39895317
|
1.966
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr19_-_47734215
|
1.962
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr3_-_9291251
|
1.922
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr11_+_19372270
|
1.860
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr11_+_10326618
|
1.824
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr14_+_23067128
|
1.815
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr5_-_112630557
|
1.741
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr1_+_45266035
|
1.739
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr15_+_65134081
|
1.703
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr5_-_149492934
|
1.702
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr8_-_28243941
|
1.700
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr1_-_11866079
|
1.688
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr5_-_112824526
|
1.666
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr19_+_6531009
|
1.632
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr10_+_72432557
|
1.630
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr14_+_70078309
|
1.604
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr6_-_39902140
|
1.590
|
NM_001075098
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr2_-_172967234
|
1.588
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr20_+_51801821
|
1.575
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr19_-_4559713
|
1.547
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr7_+_140774031
|
1.504
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chrX_-_107019001
|
1.496
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_-_43394326
|
1.472
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr8_-_95961543
|
1.465
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr3_+_183353355
|
1.455
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr19_+_55851220
|
1.428
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr6_-_11382501
|
1.424
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_+_43138919
|
1.415
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chrX_-_106959597
|
1.398
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_+_56258785
|
1.384
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr20_-_43438881
|
1.383
|
NM_001205317
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr1_-_152779106
|
1.373
|
NM_178351
|
LCE1C
|
late cornified envelope 1C
|
|
chr22_-_42828362
|
1.360
|
NM_145912
|
NFAM1
|
NFAT activating protein with ITAM motif 1
|
|
chr6_+_106546736
|
1.334
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr4_+_75230856
|
1.315
|
NM_001432
|
EREG
|
epiregulin
|
|
chr3_-_129035069
|
1.311
|
NM_006026
|
H1FX
|
H1 histone family, member X
|
|
chr16_+_11762235
|
1.309
|
NM_003498
|
SNN
|
stannin
|
|
chr12_-_123451031
|
1.297
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr19_+_41284068
|
1.296
|
NM_016154
|
RAB4B
|
RAB4B, member RAS oncogene family
|
|
chr22_+_31608249
|
1.293
|
NM_005569
|
LIMK2
|
LIM domain kinase 2
|
|
chr12_+_120427476
|
1.284
|
NM_207311
|
CCDC64
|
coiled-coil domain containing 64
|
|
chr19_-_39523132
|
1.272
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chrX_-_153775774
|
1.264
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr11_+_65554504
|
1.259
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr2_+_111878450
|
1.252
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chrX_-_153775064
|
1.247
|
NM_000402
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr5_+_121647773
|
1.239
|
NM_001242935
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr10_-_3827418
|
1.233
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr15_-_72490109
|
1.226
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr3_-_197476567
|
1.218
|
NM_001145642
|
KIAA0226
|
KIAA0226
|
|
chr6_+_106534188
|
1.213
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr12_+_117176085
|
1.207
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr22_-_30722863
|
1.203
|
NM_001204240
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr6_+_37137882
|
1.195
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr3_-_66024508
|
1.167
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr19_-_18632886
|
1.162
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr2_-_208890207
|
1.143
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr17_-_26733185
|
1.143
|
NM_001242366
NM_080669
|
SLC46A1
|
solute carrier family 46 (folate transporter), member 1
|
|
chr11_+_2466171
|
1.137
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr9_+_130854127
|
1.136
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr6_-_109330752
|
1.129
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr13_+_39261172
|
1.125
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr17_-_41985112
|
1.124
|
NM_005374
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr10_-_61900659
|
1.119
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_-_39390168
|
1.117
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr18_+_28898051
|
1.117
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr3_-_197463713
|
1.115
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr15_+_76352270
|
1.113
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr19_+_50191920
|
1.104
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr1_+_27561006
|
1.103
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr11_-_8680382
|
1.103
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr6_-_44280986
|
1.101
|
NM_020745
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr2_+_54952148
|
1.089
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr10_-_103454687
|
1.089
|
NM_022039
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr6_-_109330190
|
1.085
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr3_-_134093235
|
1.085
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr1_-_151118932
|
1.084
|
NM_001178061
NM_001178062
NM_030913
|
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C
|
|
chr5_-_111312522
|
1.075
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr9_+_130830447
|
1.058
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr9_-_117265452
|
1.053
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr3_-_36986535
|
1.048
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr5_-_111092918
|
1.041
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_-_75593720
|
1.040
|
NM_033116
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr12_+_57916607
|
1.039
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr9_+_130860816
|
1.026
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr1_-_166135951
|
1.023
|
NM_001017961
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr16_-_67217825
|
1.022
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr5_+_149997205
|
1.001
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr20_+_33814538
|
0.991
|
NM_006690
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted)
|
|
chr1_+_152784446
|
0.987
|
NM_178349
|
LCE1B
|
late cornified envelope 1B
|
|
chr1_+_204913353
|
0.984
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr16_+_67927170
|
0.979
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr5_-_111093792
|
0.978
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_46735133
|
0.974
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr10_-_62493282
|
0.959
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_+_43543877
|
0.955
|
NM_006502
|
POLH
|
polymerase (DNA directed), eta
|
|
chr21_+_40177847
|
0.952
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr17_+_7342688
|
0.944
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr1_-_59249687
|
0.938
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr14_+_61788398
|
0.936
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr12_-_53614121
|
0.935
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr15_-_37391596
|
0.928
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr9_-_117267710
|
0.925
|
NM_001173425
NM_015404
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr9_-_131534186
|
0.925
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr3_-_46718898
|
0.925
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr6_+_35420115
|
0.921
|
NM_021922
|
FANCE
|
Fanconi anemia, complementation group E
|
|
chr7_+_73245192
|
0.920
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr1_+_160370363
|
0.905
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr5_+_149980627
|
0.902
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr10_-_62332666
|
0.902
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_+_145064167
|
0.902
|
NM_000837
NM_001009184
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chrX_-_70475046
|
0.898
|
NM_001171162
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr5_-_111093251
|
0.896
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_178789597
|
0.887
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr16_-_67840464
|
0.884
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr3_+_88188261
|
0.880
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_-_169455099
|
0.879
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr12_-_53625834
|
0.869
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr14_-_91720166
|
0.858
|
NM_003485
|
GPR68
|
G protein-coupled receptor 68
|
|
chr10_-_62149487
|
0.856
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_152956563
|
0.853
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr15_-_37390372
|
0.851
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr1_-_47082529
|
0.850
|
NM_201403
|
MOB3C
|
MOB kinase activator 3C
|
|
chr9_-_130829598
|
0.848
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr15_-_74726258
|
0.845
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr5_-_111093030
|
0.840
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_+_61100653
|
0.838
|
NM_015533
|
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae)
|
|
chr14_+_24584467
|
0.823
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chrX_-_106960268
|
0.819
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr4_+_37246689
|
0.817
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr22_+_21771661
|
0.812
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr10_-_75335415
|
0.808
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr17_-_79885544
|
0.786
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr2_-_74644843
|
0.779
|
NM_001145054
|
C2orf81
|
chromosome 2 open reading frame 81
|
|
chr14_+_23305792
|
0.774
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr17_-_42201009
|
0.764
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr11_-_66115016
|
0.760
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr11_-_10715503
|
0.757
|
NM_001100163
NM_001206880
NM_001206881
NM_130385
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr15_+_41099254
|
0.755
|
NM_001077268
|
ZFYVE19
|
zinc finger, FYVE domain containing 19
|
|
chr16_-_4166185
|
0.751
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr6_-_33160199
|
0.747
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr2_+_28615771
|
0.743
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_-_151414641
|
0.742
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr3_-_13461802
|
0.740
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr4_-_142054431
|
0.739
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr14_+_24583979
|
0.713
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_33267128
|
0.710
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr2_-_73298815
|
0.709
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr6_-_109415514
|
0.708
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr12_-_50222144
|
0.704
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr16_+_57673206
|
0.699
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_-_38412725
|
0.694
|
NM_005540
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|