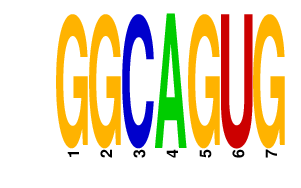
|
chrX_-_110513750
|
4.245
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr19_+_45971249
|
2.442
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr7_+_145813452
|
2.273
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr17_-_41985112
|
2.217
|
NM_005374
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr16_+_56225250
|
2.108
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr3_-_50540853
|
1.764
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr5_+_149151502
|
1.764
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr8_+_28174648
|
1.743
|
NM_006228
|
PNOC
|
prepronociceptin
|
|
chr5_+_149109814
|
1.726
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr3_-_36986535
|
1.693
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr17_-_37381920
|
1.686
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr5_+_172068188
|
1.673
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr6_+_136172802
|
1.658
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr19_+_7911385
|
1.605
|
NM_001159944
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr15_-_41166336
|
1.581
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr5_-_150948504
|
1.561
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr9_-_139440237
|
1.494
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr20_+_51588945
|
1.477
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr17_-_36762105
|
1.474
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr10_-_7453412
|
1.470
|
NM_001029880
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr2_-_122042767
|
1.465
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr20_+_51801821
|
1.456
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr19_+_7895110
|
1.388
|
NM_145245
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chrX_-_151619669
|
1.381
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr9_-_20622476
|
1.348
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr4_-_185747183
|
1.343
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr16_+_30960377
|
1.312
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr5_-_149492934
|
1.289
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr10_-_7451198
|
1.254
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr6_+_46620631
|
1.243
|
NM_001204051
NM_001204052
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr20_-_43438881
|
1.230
|
NM_001205317
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr5_-_11903964
|
1.169
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr10_-_88126201
|
1.154
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr20_-_62436855
|
1.137
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr2_+_26915390
|
1.123
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr1_+_155247186
|
1.116
|
NM_020897
|
HCN3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3
|
|
chr17_-_8066254
|
1.112
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr16_-_89007605
|
1.075
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr10_+_23983674
|
1.059
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr10_+_112631552
|
1.055
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr19_-_14316980
|
1.049
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr10_-_61900659
|
1.041
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_-_2906960
|
1.023
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr14_-_74253876
|
1.022
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr9_-_101470836
|
1.019
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr2_+_45878912
|
0.996
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr14_+_75745480
|
0.993
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr10_+_24497719
|
0.984
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr6_-_30585016
|
0.974
|
NM_002714
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10
|
|
chr11_+_117049838
|
0.972
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr14_-_89883306
|
0.962
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr12_-_13248581
|
0.953
|
NM_001206843
NM_001206845
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr17_+_29718641
|
0.944
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr12_-_56652118
|
0.943
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr19_-_49016418
|
0.941
|
NM_001080434
|
LMTK3
|
lemur tyrosine kinase 3
|
|
chr10_+_5566923
|
0.935
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr1_-_22469414
|
0.927
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr14_-_90085325
|
0.916
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr1_+_3607232
|
0.915
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chr9_-_89562103
|
0.908
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr14_-_99737821
|
0.902
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr14_-_74226890
|
0.891
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr18_+_55862577
|
0.888
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr15_+_90744506
|
0.878
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr15_+_48010685
|
0.858
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr18_+_55714457
|
0.854
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr22_-_38577692
|
0.849
|
NM_001004426
NM_003560
NM_001199562
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr18_+_55711388
|
0.834
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_109605668
|
0.830
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr1_+_204485461
|
0.828
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr18_+_55816546
|
0.823
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_151414641
|
0.816
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr22_+_35695267
|
0.815
|
NM_001135729
|
TOM1
|
target of myb1 (chicken)
|
|
chr16_-_89043215
|
0.815
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr12_+_10331556
|
0.813
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr2_-_239197091
|
0.810
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr22_+_40573879
|
0.808
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr20_-_62168663
|
0.805
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr1_-_41950296
|
0.802
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr11_-_8190535
|
0.792
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr10_-_135238075
|
0.791
|
NM_001012508
|
SPRN
|
shadow of prion protein homolog (zebrafish)
|
|
chr1_+_160085519
|
0.789
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr22_+_35695792
|
0.786
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr11_+_537521
|
0.785
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr20_+_37434294
|
0.785
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr10_-_62493282
|
0.780
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_3569098
|
0.779
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr1_+_11796128
|
0.779
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr11_+_19734821
|
0.769
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr16_+_87636492
|
0.754
|
NM_020655
|
JPH3
|
junctophilin 3
|
|
chr19_+_41082729
|
0.747
|
NM_138392
|
SHKBP1
|
SH3KBP1 binding protein 1
|
|
chr16_+_22217369
|
0.744
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr3_-_18466759
|
0.744
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr12_-_13256572
|
0.741
|
NM_001080554
NM_001080555
NM_001206842
|
GSG1
|
germ cell associated 1
|
|
chr10_-_62149487
|
0.741
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_-_63536112
|
0.735
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr19_-_47617008
|
0.731
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr12_+_57916607
|
0.731
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr11_+_19372270
|
0.725
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr18_+_55888750
|
0.725
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_65877309
|
0.715
|
NM_004480
NM_178155
NM_178156
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr11_-_67442086
|
0.708
|
NM_000695
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr17_-_27224708
|
0.703
|
NM_004475
|
FLOT2
|
flotillin 2
|
|
chr5_-_131826426
|
0.692
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr11_-_67448684
|
0.688
|
NM_001031615
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr19_+_18794391
|
0.685
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr12_+_107712151
|
0.682
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr9_+_131799212
|
0.679
|
NM_032809
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr10_-_64576123
|
0.674
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr12_-_125052009
|
0.674
|
NM_001077261
NM_001206654
NM_006312
|
NCOR2
|
nuclear receptor corepressor 2
|
|
chr3_+_39851028
|
0.672
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr17_+_43971655
|
0.661
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr1_-_112298418
|
0.644
|
NM_198926
|
FAM212B
|
family with sequence similarity 212, member B
|
|
chr22_+_40440664
|
0.643
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_+_3614609
|
0.642
|
NM_001204192
|
TP73
|
tumor protein p73
|
|
chr15_+_90728118
|
0.641
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr15_+_47476225
|
0.635
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr10_+_1102741
|
0.630
|
NM_014023
|
WDR37
|
WD repeat domain 37
|
|
chr11_+_3829765
|
0.623
|
NM_014489
|
PGAP2
|
post-GPI attachment to proteins 2
|
|
chr10_-_62332666
|
0.621
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_-_18480203
|
0.620
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr3_-_141747434
|
0.619
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr2_-_157189040
|
0.616
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chrX_+_105969875
|
0.615
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr15_+_79724857
|
0.613
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr3_+_25469833
|
0.610
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr17_-_48227873
|
0.609
|
NM_032595
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B
|
|
chr11_+_92085261
|
0.602
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr7_-_100286869
|
0.596
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr10_-_64578926
|
0.591
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr11_-_66488712
|
0.586
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr15_-_40398573
|
0.585
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr8_-_60031545
|
0.582
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr11_+_3818953
|
0.582
|
NM_001145438
|
PGAP2
|
post-GPI attachment to proteins 2
|
|
chr1_+_66258192
|
0.582
|
NM_001037341
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr11_+_60681370
|
0.581
|
NM_024092
|
TMEM109
|
transmembrane protein 109
|
|
chr1_+_66258855
|
0.579
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr5_+_133861685
|
0.578
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr5_-_175964223
|
0.575
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr19_-_44143943
|
0.569
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr20_+_62371109
|
0.563
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr8_+_102504661
|
0.560
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr3_-_141868208
|
0.559
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr5_+_149887673
|
0.555
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr16_+_29817810
|
0.549
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr10_-_105677986
|
0.546
|
NM_024928
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1
|
|
chr10_-_94050799
|
0.545
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr18_+_55019720
|
0.545
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr2_+_166428845
|
0.543
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr2_+_166326156
|
0.543
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr5_-_137071703
|
0.542
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr7_+_138916230
|
0.541
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr15_-_40401041
|
0.538
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr2_-_220094354
|
0.537
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chr15_-_40398286
|
0.535
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr15_+_32322690
|
0.534
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr9_-_135996287
|
0.531
|
NM_006266
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr1_-_112281864
|
0.529
|
NM_019099
|
FAM212B
|
family with sequence similarity 212, member B
|
|
chr21_-_34100316
|
0.526
|
NM_003895
NM_203446
|
SYNJ1
|
synaptojanin 1
|
|
chr1_-_154842725
|
0.523
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr11_-_46722115
|
0.518
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr17_-_1928177
|
0.516
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr1_-_161015752
|
0.515
|
NM_007122
NM_207005
|
USF1
|
upstream transcription factor 1
|
|
chr21_-_34100228
|
0.514
|
NM_001160302
NM_001160306
|
SYNJ1
|
synaptojanin 1
|
|
chr12_-_2986299
|
0.514
|
NM_001243088
NM_001243089
NM_021953
NM_202002
NM_202003
|
FOXM1
|
forkhead box M1
|
|
chr5_-_134914968
|
0.511
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr18_+_7567313
|
0.509
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chrX_+_152990322
|
0.509
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr1_+_6484816
|
0.508
|
NM_031475
|
ESPN
|
espin
|
|
chr11_+_64794879
|
0.505
|
NM_013306
NM_147777
|
SNX15
|
sorting nexin 15
|
|
chr11_-_118047336
|
0.505
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr9_-_131534186
|
0.500
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr22_-_37545944
|
0.498
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr16_-_31519629
|
0.498
|
NM_022744
|
C16orf58
|
chromosome 16 open reading frame 58
|
|
chr3_-_123135188
|
0.498
|
NM_001199642
|
ADCY5
|
adenylate cyclase 5
|
|
chr1_-_151431892
|
0.497
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr1_+_66797775
|
0.497
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chrX_+_9983793
|
0.491
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chrX_+_147582133
|
0.485
|
NM_001169122
NM_001169123
NM_001169124
NM_001169125
NM_002025
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr1_+_11539177
|
0.485
|
NM_020780
|
PTCHD2
|
patched domain containing 2
|
|
chr8_+_77593506
|
0.479
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr9_+_131549509
|
0.479
|
NM_018201
|
TBC1D13
|
TBC1 domain family, member 13
|
|
chr11_+_61447816
|
0.478
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr2_-_200335988
|
0.476
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr17_+_2240795
|
0.475
|
NM_014853
NM_001098509
|
SGSM2
|
small G protein signaling modulator 2
|
|
chr6_-_82462374
|
0.474
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr1_+_66458389
|
0.474
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr19_-_46295785
|
0.469
|
NM_004943
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr14_+_68168602
|
0.467
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr9_-_136024522
|
0.460
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr1_+_184356128
|
0.460
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr10_-_94003002
|
0.459
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr19_+_10713055
|
0.459
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr5_+_6448735
|
0.455
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr15_-_101084871
|
0.454
|
NM_178842
|
CERS3
|
ceramide synthase 3
|
|
chr16_+_30064410
|
0.453
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr3_+_25469749
|
0.451
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr5_-_160279045
|
0.445
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr9_-_37576248
|
0.445
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr3_-_141719188
|
0.443
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|