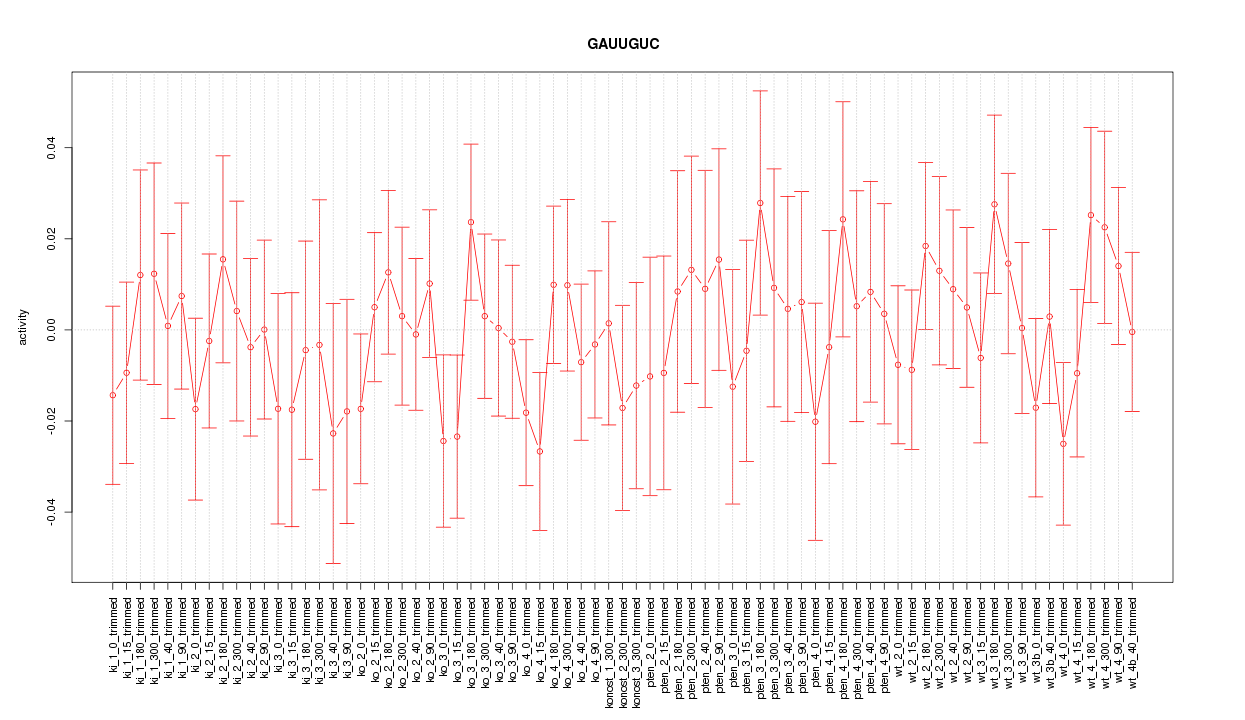
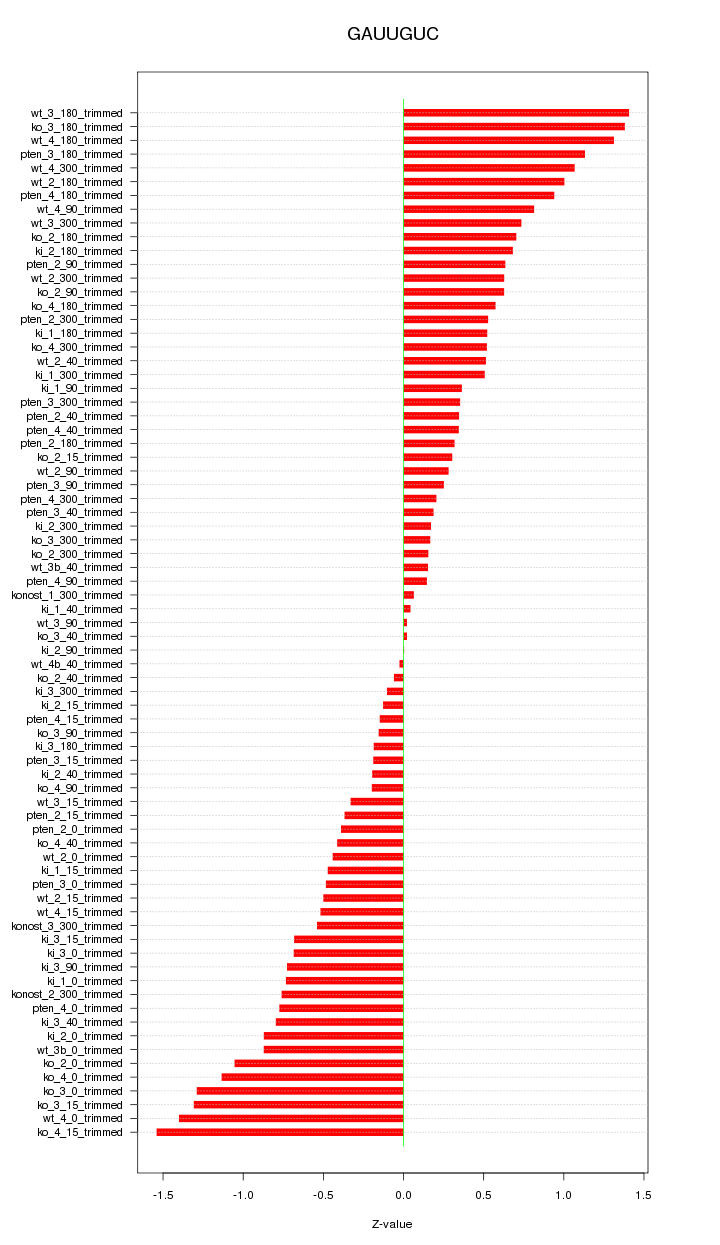
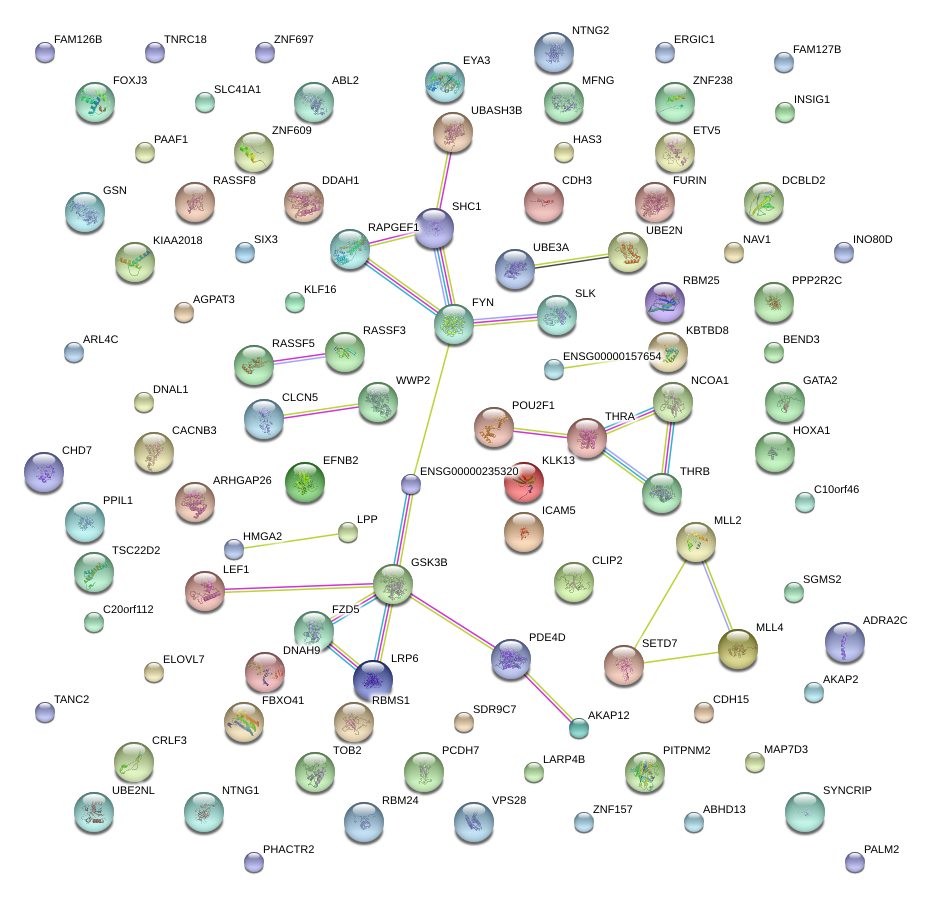
|
chr6_+_151646634
|
7.606
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_151561094
|
7.007
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_+_107682628
|
2.555
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr11_+_122526339
|
2.458
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr3_+_67048726
|
2.231
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr16_+_69139466
|
2.185
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr16_+_69141442
|
2.155
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_-_205782135
|
2.021
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr1_-_86043932
|
1.935
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_-_98620452
|
1.928
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr12_+_65004191
|
1.927
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr5_+_142150291
|
1.914
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr5_-_60140041
|
1.895
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL fatty acid elongase 7
|
|
chr3_-_128206704
|
1.830
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr15_+_91411884
|
1.811
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr3_-_128212027
|
1.714
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr2_-_208634051
|
1.700
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr15_+_64791618
|
1.692
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chr2_+_45169036
|
1.642
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr17_-_29151724
|
1.618
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr5_-_58882274
|
1.608
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_128207366
|
1.601
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr13_-_107187312
|
1.593
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr6_-_107435635
|
1.583
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr3_-_185826748
|
1.515
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr19_-_1863546
|
1.509
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr1_-_154943001
|
1.502
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr1_-_120190389
|
1.481
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr2_+_64681270
|
1.478
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr1_+_244216506
|
1.451
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chrX_+_49832214
|
1.426
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_-_85930733
|
1.395
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_-_59783885
|
1.378
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_66218141
|
1.368
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr7_+_73703704
|
1.364
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr5_-_58571916
|
1.360
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_-_41842853
|
1.358
|
NM_016272
|
TOB2
|
transducer of ERBB2, 2
|
|
chr12_-_123594974
|
1.348
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr4_-_109089444
|
1.337
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr8_+_61591239
|
1.323
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr4_+_30721865
|
1.272
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr2_-_235405692
|
1.240
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr12_+_49209300
|
1.225
|
NM_001206916
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr12_+_49208214
|
1.224
|
NM_001206917
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr4_+_3768295
|
1.215
|
NM_000683
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr2_-_161350304
|
1.197
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr20_-_31071187
|
1.180
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr17_+_61086897
|
1.175
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr3_-_24536176
|
1.171
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr1_-_42801529
|
1.162
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chrX_+_49687212
|
1.127
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr7_-_27135524
|
1.127
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr6_-_86352635
|
1.126
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr5_-_58334936
|
1.101
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_201936389
|
1.100
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr12_-_12419702
|
1.099
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr12_+_26205490
|
1.097
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_+_244214552
|
1.074
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr12_+_26126687
|
1.069
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_+_167298280
|
1.061
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr15_-_25650647
|
1.046
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr1_-_179112178
|
1.036
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr4_+_108814419
|
0.993
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr6_-_86352564
|
0.989
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr3_+_150126705
|
0.978
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chrX_-_135338640
|
0.967
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr12_+_49212253
|
0.967
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr6_-_86351156
|
0.954
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr5_-_59189620
|
0.951
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_93835986
|
0.949
|
NM_003348
|
UBE2N
|
ubiquitin-conjugating enzyme E2N
|
|
chr5_-_58295758
|
0.944
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_-_37882214
|
0.940
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr3_+_187943192
|
0.925
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_-_119812973
|
0.923
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr7_-_5463176
|
0.887
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr4_+_108745718
|
0.886
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr14_+_74111577
|
0.881
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr6_+_143929316
|
0.879
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr14_+_73525187
|
0.860
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr9_+_112542496
|
0.854
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr5_+_172261220
|
0.854
|
NM_001031711
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr5_-_58652671
|
0.854
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_6383572
|
0.839
|
NM_181876
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr6_-_36842799
|
0.827
|
NM_016059
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1
|
|
chr10_-_931694
|
0.825
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr1_-_42800883
|
0.823
|
NM_001198852
|
FOXJ3
|
forkhead box J3
|
|
chr21_+_45345278
|
0.823
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr2_+_198380294
|
0.821
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr6_+_143999058
|
0.788
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_-_28415071
|
0.787
|
NM_001990
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr16_+_68678150
|
0.786
|
NM_001793
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr10_+_105727469
|
0.779
|
NM_014720
|
SLK
|
STE20-like kinase
|
|
chr2_-_206950895
|
0.777
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr16_+_69958900
|
0.777
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr1_+_201617449
|
0.767
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr1_-_179198814
|
0.766
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr4_-_6422844
|
0.751
|
NM_001206996
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr6_+_17282931
|
0.751
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr9_-_134612924
|
0.749
|
NM_005312
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr4_-_109087876
|
0.746
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr2_+_24807345
|
0.739
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr12_-_49449106
|
0.737
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr9_+_112403067
|
0.727
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr4_-_140477290
|
0.717
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr1_-_42800623
|
0.715
|
NM_001198851
|
FOXJ3
|
forkhead box J3
|
|
chr2_-_73496595
|
0.712
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr1_-_42800902
|
0.705
|
NM_014947
|
FOXJ3
|
forkhead box J3
|
|
chr10_-_120514669
|
0.697
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr1_-_154946859
|
0.696
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_+_155089483
|
0.689
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr4_-_6557404
|
0.688
|
NM_001206995
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr13_+_108870713
|
0.683
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr12_-_109125289
|
0.680
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr5_+_150827138
|
0.668
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr6_-_16761600
|
0.633
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr1_+_29562947
|
0.633
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr8_+_23101149
|
0.632
|
NM_152272
|
CHMP7
|
charged multivesicular body protein 7
|
|
chr14_+_55518313
|
0.627
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr18_+_43914186
|
0.622
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr13_-_26795821
|
0.619
|
NM_183044
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr1_+_160336856
|
0.617
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr17_-_37764165
|
0.602
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr3_-_170626425
|
0.597
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr8_+_104033241
|
0.594
|
NM_001695
|
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1
|
|
chr3_+_30647901
|
0.587
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr8_-_133459508
|
0.582
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr6_+_17281773
|
0.578
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_65659634
|
0.577
|
NM_181784
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr2_-_148779172
|
0.571
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr2_+_198380752
|
0.568
|
NM_001100819
NM_015387
|
MOB4
|
MOB family member 4, phocein
|
|
chr2_+_198381021
|
0.563
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr2_-_65593866
|
0.559
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr13_-_26795927
|
0.558
|
NM_183043
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr19_-_3700466
|
0.540
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr13_-_26796343
|
0.539
|
NM_005977
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr1_+_201708940
|
0.539
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr2_-_204399906
|
0.533
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr9_+_115983807
|
0.527
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr1_-_212004113
|
0.518
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr6_-_86352982
|
0.514
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_199996729
|
0.514
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr16_+_57126439
|
0.512
|
NM_152727
|
CPNE2
|
copine II
|
|
chr4_-_152147520
|
0.511
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr8_-_81787015
|
0.502
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr7_-_127032717
|
0.501
|
NM_176814
|
ZNF800
|
zinc finger protein 800
|
|
chr4_-_6565326
|
0.497
|
NM_001206994
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr3_-_183145854
|
0.496
|
NM_015078
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2
|
|
chr14_-_93799371
|
0.495
|
NM_001002860
NM_018167
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr2_-_70994846
|
0.489
|
NM_001185054
|
ADD2
|
adducin 2 (beta)
|
|
chr2_+_101436491
|
0.488
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr4_+_48343502
|
0.486
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr10_+_11206928
|
0.484
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_-_132299263
|
0.481
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr3_+_185000714
|
0.470
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr7_-_131241321
|
0.467
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr11_-_27528298
|
0.464
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chrX_+_128674181
|
0.461
|
NM_000276
NM_001587
|
OCRL
|
oculocerebrorenal syndrome of Lowe
|
|
chrX_+_103411065
|
0.457
|
NM_207318
|
FAM199X
|
family with sequence similarity 199, X-linked
|
|
chr20_+_4666796
|
0.456
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr3_-_125094003
|
0.455
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr9_-_134585107
|
0.454
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr8_-_133493003
|
0.448
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr12_+_26111951
|
0.448
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr11_-_18655962
|
0.448
|
NM_194285
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr4_+_75858205
|
0.443
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr18_+_158480
|
0.434
|
NM_001037334
NM_005151
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr6_+_47445440
|
0.433
|
NM_012120
|
CD2AP
|
CD2-associated protein
|
|
chr4_-_152096655
|
0.416
|
NM_001128924
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_-_14871555
|
0.406
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr3_+_43328001
|
0.405
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr19_+_1285766
|
0.402
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr2_+_74273449
|
0.394
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr15_+_85923739
|
0.387
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr12_-_120554612
|
0.371
|
NM_001167606
NM_006861
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr1_+_171454636
|
0.366
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr17_+_77751929
|
0.363
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr13_+_100258886
|
0.363
|
NM_206808
|
CLYBL
|
citrate lyase beta like
|
|
chr12_-_56652118
|
0.360
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr14_-_73925279
|
0.356
|
NM_001005743
NM_001005744
NM_001005745
NM_003744
|
NUMB
|
numb homolog (Drosophila)
|
|
chr15_+_86220267
|
0.350
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr1_-_155532322
|
0.350
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr10_-_104953010
|
0.349
|
NM_001134373
NM_012229
|
NT5C2
|
5'-nucleotidase, cytosolic II
|
|
chr1_-_31230673
|
0.349
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr9_+_102584136
|
0.346
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr9_+_110045516
|
0.344
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr17_+_31254927
|
0.343
|
NM_001033504
NM_015544
|
TMEM98
|
transmembrane protein 98
|
|
chr1_+_65730423
|
0.333
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr7_+_21467572
|
0.324
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr5_-_114598547
|
0.323
|
NM_005023
|
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit
|
|
chr2_-_69614321
|
0.309
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr12_-_42538432
|
0.305
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr1_-_31381436
|
0.304
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr9_+_95087740
|
0.302
|
NM_001012267
|
CENPP
|
centromere protein P
|
|
chr17_+_80477564
|
0.297
|
NM_004514
|
FOXK2
|
forkhead box K2
|
|
chr14_+_55221505
|
0.294
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_156470506
|
0.280
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr19_-_2051222
|
0.278
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr14_+_71996041
|
0.278
|
NM_015556
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1
|
|
chr15_-_25684161
|
0.275
|
NM_000462
NM_130839
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chrX_-_135333519
|
0.271
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|