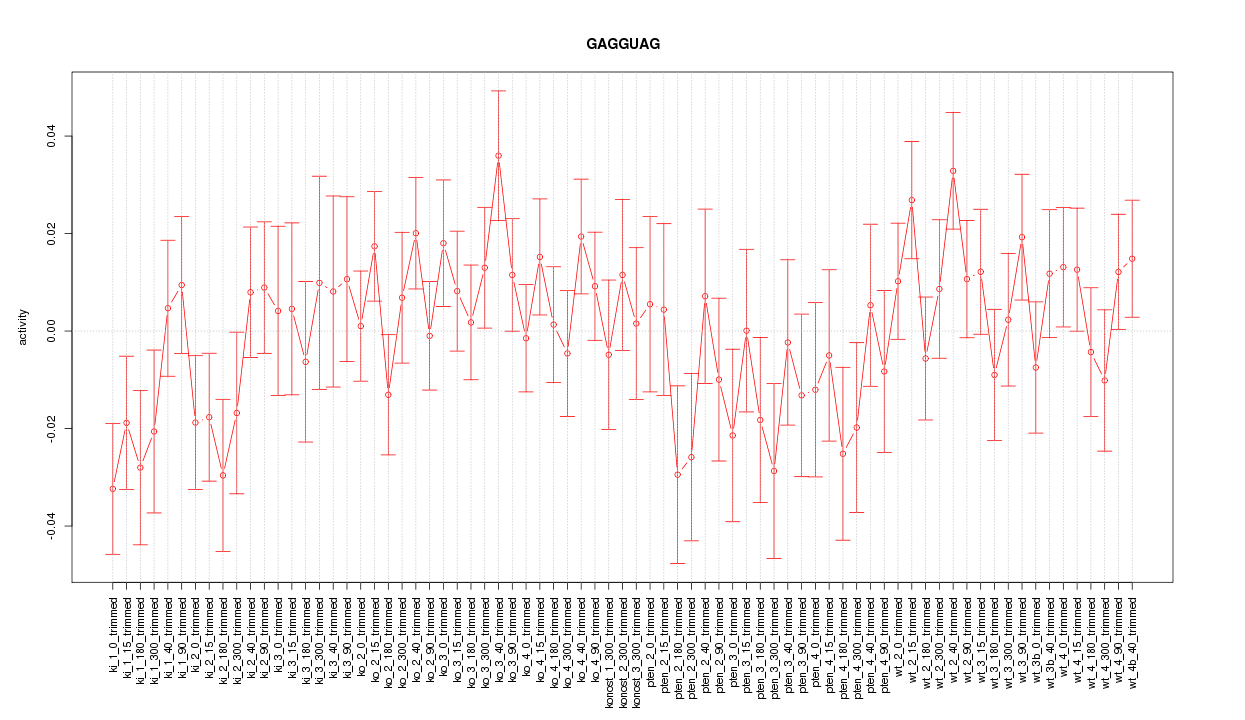
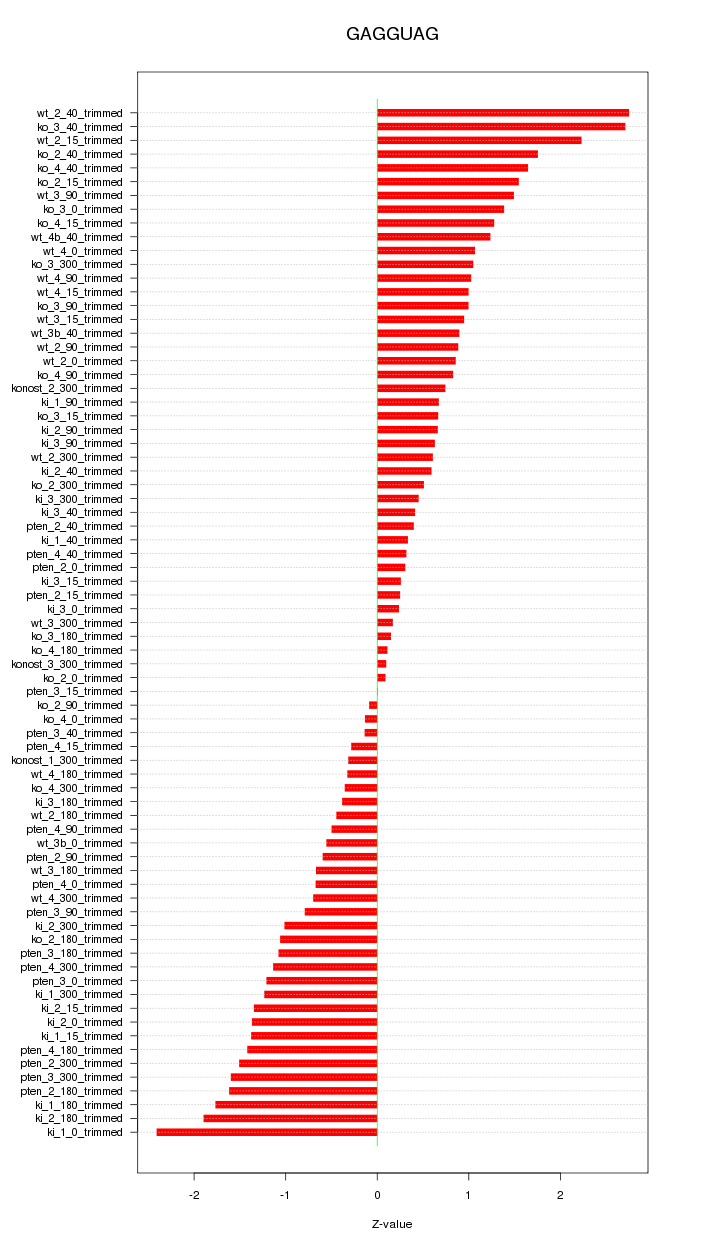
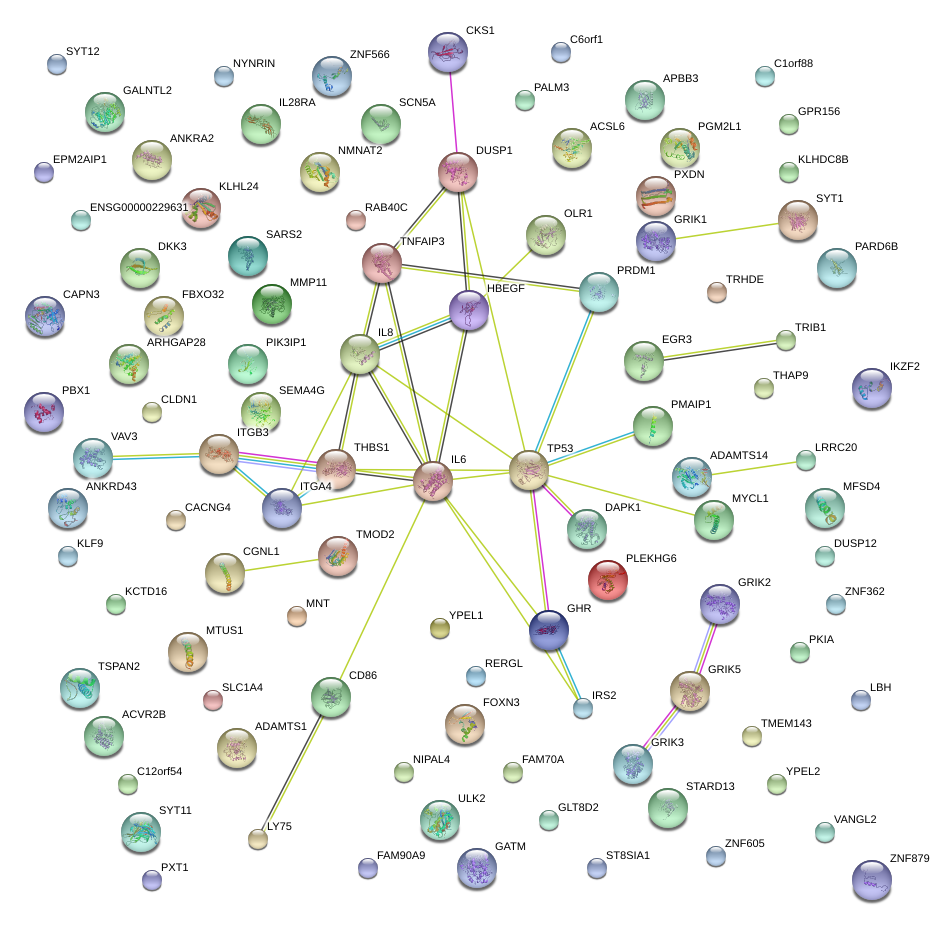
|
chr2_+_30454396
|
10.713
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr5_+_156887026
|
8.339
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr8_-_124544376
|
8.032
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr8_-_124553448
|
7.849
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr8_-_22549901
|
7.406
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
7.366
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr1_-_115632047
|
7.358
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr8_-_22550708
|
7.212
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr22_-_31688329
|
6.684
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr3_+_49208979
|
6.575
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr14_+_24867926
|
6.385
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr7_+_22766765
|
6.188
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr5_+_178450775
|
6.052
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr4_+_74606222
|
5.820
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr9_+_90112754
|
5.448
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr2_-_1748285
|
5.097
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr11_-_12030128
|
4.821
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr18_+_6834431
|
4.629
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr13_-_110438896
|
4.460
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr17_-_19770988
|
4.396
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr6_+_138188352
|
4.228
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_-_24513737
|
4.144
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr15_-_45670955
|
4.115
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr15_+_57668702
|
4.037
|
NM_001252335
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr22_+_24115035
|
4.028
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr5_+_42423811
|
4.013
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr6_+_106546736
|
3.962
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_+_42424553
|
3.907
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr12_+_6419601
|
3.786
|
NM_018173
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr6_+_106534188
|
3.765
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr18_+_57567191
|
3.746
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr15_+_52043738
|
3.742
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr5_-_131347518
|
3.648
|
NM_001205248
NM_001205251
NM_001205250
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr1_-_108507474
|
3.617
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr2_+_65216482
|
3.611
|
NM_003038
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr1_-_40367539
|
3.606
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr12_-_133532847
|
3.599
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr5_+_42425093
|
3.522
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr3_-_38691118
|
3.509
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr9_-_73029539
|
3.432
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr5_+_42548144
|
3.376
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr12_+_72666462
|
3.366
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr3_+_183353355
|
3.366
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr5_+_42565963
|
3.356
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr5_+_42548314
|
3.352
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr15_+_42651697
|
3.333
|
NM_000070
NM_024344
NM_173087
|
CAPN3
|
calpain 3, (p94)
|
|
chr1_+_111889194
|
3.301
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr19_-_36980336
|
3.240
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr1_-_183387501
|
3.237
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr5_+_42549657
|
3.186
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr19_-_14169970
|
3.176
|
NM_001145028
|
PALM3
|
paralemmin 3
|
|
chr5_-_139726173
|
3.124
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr8_+_126442557
|
3.100
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr5_+_42550181
|
3.072
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr3_-_119963141
|
3.054
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr15_+_42696957
|
3.050
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr5_+_132149022
|
2.950
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr5_-_131347249
|
2.944
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr17_+_57408907
|
2.913
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr10_-_72142344
|
2.896
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr2_+_65215577
|
2.890
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr1_-_108231122
|
2.885
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr10_-_72141347
|
2.877
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr1_-_183274008
|
2.787
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr1_+_164528586
|
2.777
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr15_+_39873276
|
2.777
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr3_+_16216155
|
2.730
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr5_+_42565562
|
2.716
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr1_+_155829259
|
2.698
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr17_+_45331154
|
2.629
|
NM_000212
|
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61)
|
|
chr15_+_42694587
|
2.612
|
NM_173088
|
CAPN3
|
calpain 3, (p94)
|
|
chr10_+_102732285
|
2.592
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr11_-_12030882
|
2.576
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_+_205538111
|
2.570
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr5_-_139944045
|
2.568
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr12_+_6420098
|
2.560
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr20_+_49348050
|
2.541
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr5_-_72861376
|
2.536
|
NM_023039
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr1_+_160370363
|
2.428
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr5_+_143550395
|
2.418
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr11_-_12030622
|
2.414
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr3_+_121774208
|
2.376
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chrX_-_119445306
|
2.286
|
NM_001104544
NM_001104545
NM_017938
|
FAM70A
|
family with sequence similarity 70, member A
|
|
chr1_+_33722158
|
2.285
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr8_+_79428335
|
2.277
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr2_-_160761266
|
2.256
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr12_+_6421816
|
2.250
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr8_-_17658385
|
2.235
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_36410665
|
2.230
|
NM_152990
|
PXT1
|
peroxisomal, testis specific 1
|
|
chr6_+_101846860
|
2.229
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr10_+_72432557
|
2.215
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr2_+_182321618
|
2.188
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr3_+_38495778
|
2.106
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr12_+_79258448
|
2.100
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79257772
|
2.091
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr4_+_83821835
|
2.072
|
NM_024672
|
THAP9
|
THAP domain containing 9
|
|
chr14_-_90085325
|
2.070
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr12_-_22487566
|
2.060
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr12_+_79439432
|
2.053
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr5_-_172198160
|
2.024
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr3_+_121796695
|
1.978
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr12_-_10324675
|
1.960
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr3_-_190040192
|
1.951
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr16_+_639668
|
1.936
|
NM_001172664
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr17_-_7590731
|
1.935
|
NM_000546
NM_001126112
NM_001126113
NM_001126114
|
TP53
|
tumor protein p53
|
|
chr21_-_28217692
|
1.913
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr13_-_33760215
|
1.901
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_214016332
|
1.893
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr17_+_64960754
|
1.875
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr16_+_640056
|
1.875
|
NM_001172665
NM_001172666
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr19_-_48867149
|
1.851
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr17_-_2304217
|
1.829
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr11_-_74109417
|
1.819
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr8_-_122653493
|
1.798
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr14_-_89883306
|
1.785
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr9_+_123850573
|
1.779
|
NM_007018
|
CNTRL
|
centriolin
|
|
chr12_-_111021026
|
1.776
|
NM_139283
|
PPTC7
|
PTC7 protein phosphatase homolog (S. cerevisiae)
|
|
chr13_-_33859835
|
1.766
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_7578810
|
1.765
|
NM_001126115
NM_001126116
NM_001126117
|
TP53
|
tumor protein p53
|
|
chr3_+_102153858
|
1.755
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr13_-_96296919
|
1.739
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr2_-_180129245
|
1.736
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr13_+_39612440
|
1.727
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr16_+_639356
|
1.684
|
NM_001172663
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr3_-_132441302
|
1.683
|
NM_153240
|
NPHP3-ACAD11
NPHP3
|
NPHP3-ACAD11 readthrough
nephronophthisis 3 (adolescent)
|
|
chr21_+_30671217
|
1.682
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr14_+_45366426
|
1.676
|
NM_001017923
|
C14orf28
|
chromosome 14 open reading frame 28
|
|
chr8_-_95961543
|
1.665
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr17_-_38721701
|
1.658
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr11_-_74204708
|
1.636
|
NM_001144869
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative)
|
|
chr3_-_87040246
|
1.631
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_-_185126153
|
1.603
|
NM_001202423
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr3_-_69435429
|
1.579
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr3_+_50712671
|
1.565
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr1_-_185125819
|
1.564
|
NM_030934
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr9_-_77502635
|
1.563
|
NM_001177310
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr1_+_213123853
|
1.554
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr16_+_640172
|
1.545
|
NM_021168
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr21_+_27107257
|
1.537
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr6_+_12290528
|
1.515
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr7_-_20826503
|
1.508
|
NM_182700
NM_198956
|
SP8
|
Sp8 transcription factor
|
|
chr7_-_6591066
|
1.505
|
NM_001145118
|
GRID2IP
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein
|
|
chr3_-_9291251
|
1.491
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr11_-_27494232
|
1.473
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr7_-_112579726
|
1.473
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr20_-_5591487
|
1.467
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr8_-_109095836
|
1.449
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chr19_-_7293876
|
1.427
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr21_+_30671736
|
1.403
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chrX_-_54384436
|
1.402
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr13_-_33780142
|
1.392
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_49460110
|
1.383
|
NM_000481
NM_001164710
NM_001164711
NM_001164712
|
AMT
|
aminomethyltransferase
|
|
chr7_+_20370724
|
1.381
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr4_+_15004297
|
1.372
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr16_+_28303622
|
1.337
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr21_+_27107322
|
1.314
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr9_+_4662285
|
1.307
|
NM_203453
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr10_-_94050799
|
1.304
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_+_107241782
|
1.291
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr18_+_11751470
|
1.290
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr12_-_95611023
|
1.282
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr10_-_61469322
|
1.250
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr17_-_61777478
|
1.249
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr2_+_74881353
|
1.248
|
NM_004263
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F
|
|
chr9_+_19408924
|
1.247
|
NM_001010887
|
ACER2
|
alkaline ceramidase 2
|
|
chr20_-_30795346
|
1.245
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr6_-_134373618
|
1.238
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr14_+_53019865
|
1.235
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr12_-_69326946
|
1.233
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr3_-_36986535
|
1.225
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr19_+_6531009
|
1.220
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr3_+_53529030
|
1.217
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr13_+_39261172
|
1.207
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr11_-_16430425
|
1.204
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr5_-_131132715
|
1.204
|
NM_001008738
NM_133372
|
FNIP1
|
folliculin interacting protein 1
|
|
chr12_-_54673901
|
1.198
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr1_-_92371558
|
1.197
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr3_-_143567163
|
1.192
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr10_-_94003002
|
1.185
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_16870719
|
1.180
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr5_+_72921980
|
1.174
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr8_+_104383785
|
1.161
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr5_-_122759003
|
1.159
|
NM_001166226
NM_153223
|
CEP120
|
centrosomal protein 120kDa
|
|
chr3_+_112051914
|
1.153
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr3_+_39851028
|
1.146
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr1_+_185126191
|
1.140
|
NM_001105518
NM_017673
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae)
|
|
chr1_+_36273786
|
1.130
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr11_-_16497917
|
1.099
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr17_+_34058678
|
1.099
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr17_-_65241305
|
1.090
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr15_+_99192696
|
1.088
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr5_+_150157507
|
1.083
|
NM_032947
|
C5orf62
|
chromosome 5 open reading frame 62
|
|
chr18_+_21032786
|
1.081
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr1_-_92351613
|
1.075
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_203274646
|
1.074
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr8_-_42751622
|
1.065
|
NM_001160223
NM_001160224
NM_001160225
NM_030954
|
RNF170
|
ring finger protein 170
|
|
chr2_+_86668270
|
1.046
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr12_-_123451031
|
1.032
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr9_+_131182690
|
1.029
|
NM_016174
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr20_+_37434294
|
1.028
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|