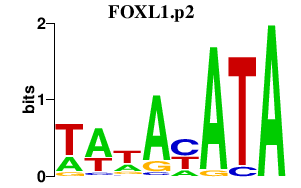
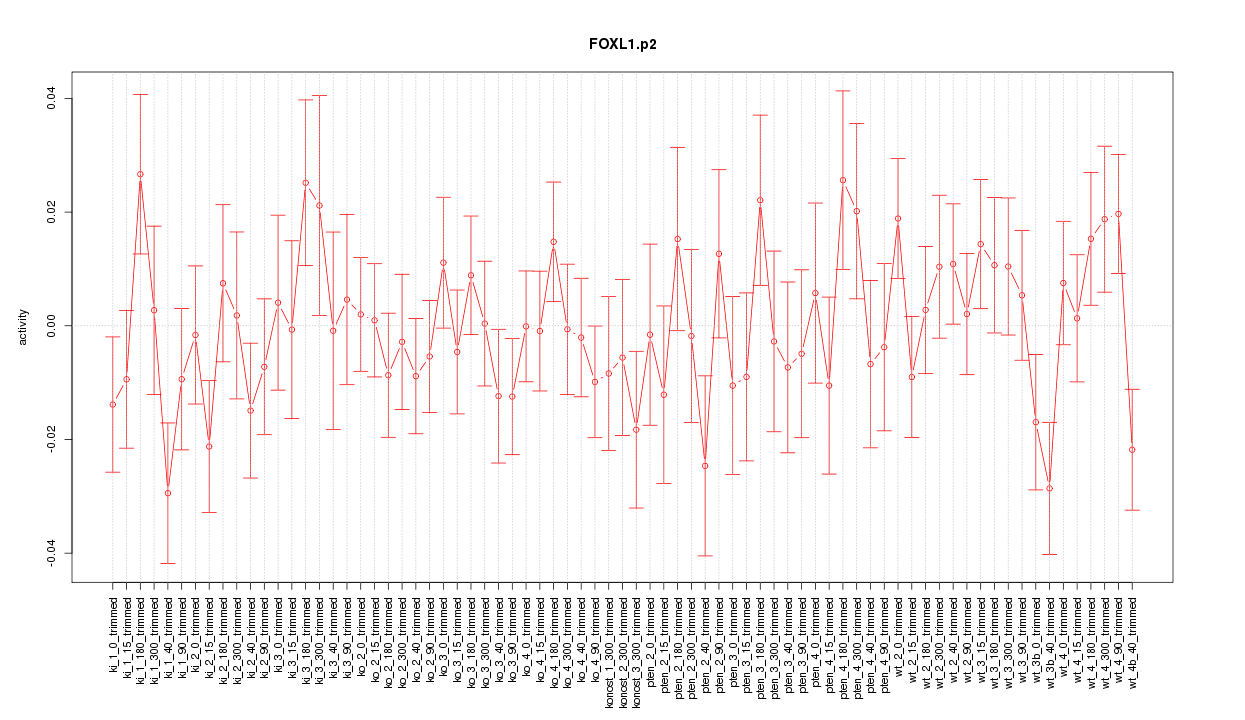
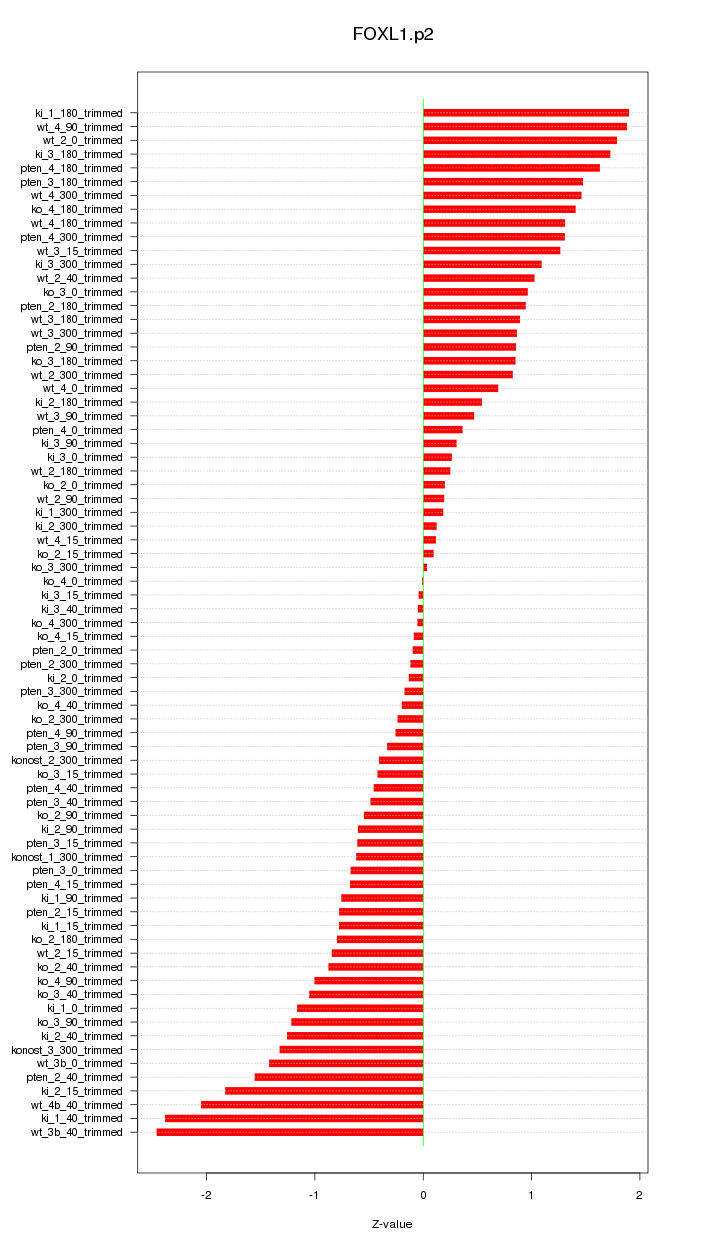
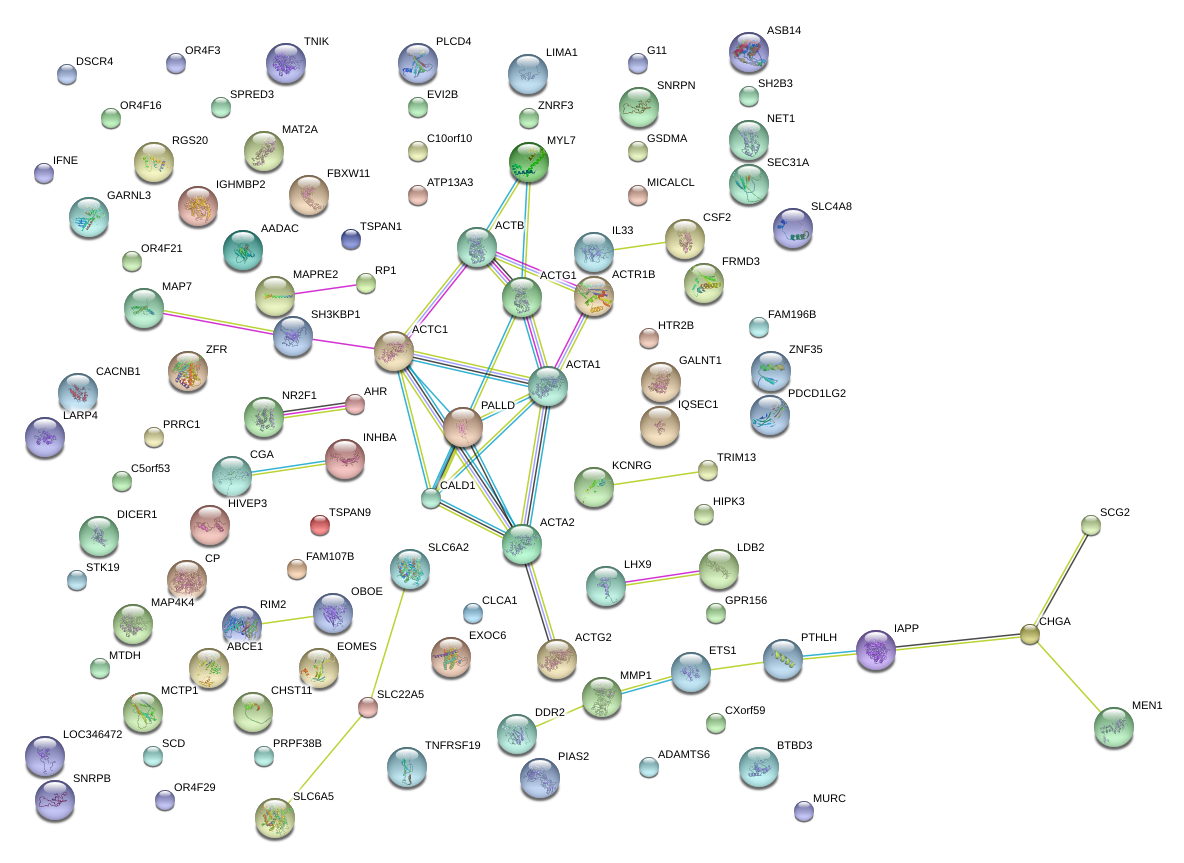
|
chr12_+_50869531
|
7.531
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr3_-_12941547
|
6.255
|
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr13_+_50589389
|
4.585
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr11_+_12308446
|
4.527
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr8_+_104831415
|
3.834
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr19_+_39899749
|
3.786
|
|
|
|
|
chr17_-_29641087
|
3.681
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_29641094
|
3.630
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_-_42384370
|
3.566
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr9_+_5510544
|
3.477
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr3_+_44692500
|
3.215
|
|
ZNF35
|
zinc finger protein 35
|
|
chr5_-_94224601
|
3.132
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr1_+_367639
|
3.048
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr3_-_194188967
|
2.978
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr14_-_95599794
|
2.957
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr7_-_44180911
|
2.902
|
NM_021223
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr7_-_41742666
|
2.856
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr5_+_139505519
|
2.753
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr17_-_29641068
|
2.714
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_-_32388687
|
2.687
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr2_-_55199894
|
2.659
|
|
|
|
|
chr4_+_169418167
|
2.648
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr1_-_622033
|
2.560
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr9_-_85882093
|
2.473
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr11_-_102668822
|
2.448
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr10_-_45474210
|
2.368
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr12_-_28124915
|
2.368
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr1_+_162602198
|
2.350
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr9_+_6215785
|
2.344
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr5_-_94417185
|
2.332
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr4_-_16900028
|
2.309
|
|
LDB2
|
LIM domain binding 2
|
|
chr8_+_54764367
|
2.252
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr10_+_5488521
|
2.246
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr18_+_33234532
|
2.139
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr9_-_21482311
|
2.118
|
NM_176891
|
IFNE
|
interferon, epsilon
|
|
chr2_+_102508377
|
2.088
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_+_134464161
|
2.087
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr7_+_17382900
|
2.047
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chr7_+_134464417
|
1.986
|
|
CALD1
|
caldesmon 1
|
|
chr3_-_27764197
|
1.951
|
|
EOMES
|
eomesodermin
|
|
chr19_+_38880839
|
1.926
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr10_+_102121371
|
1.921
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr12_+_105153258
|
1.916
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr17_+_38119225
|
1.876
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr9_+_130026755
|
1.863
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr3_-_119963141
|
1.859
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr12_+_65224214
|
1.857
|
|
|
|
|
chr5_-_169407743
|
1.793
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr3_-_57326709
|
1.771
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr10_-_21807120
|
1.737
|
|
|
|
|
chr2_-_224467049
|
1.715
|
|
SCG2
|
secretogranin II
|
|
chr4_+_146031528
|
1.691
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr7_+_134464372
|
1.691
|
|
CALD1
|
caldesmon 1
|
|
chr5_+_86676620
|
1.660
|
|
|
|
|
chr3_-_148939784
|
1.655
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr5_+_126859157
|
1.648
|
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr10_+_94594469
|
1.593
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr11_-_102668892
|
1.543
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr6_-_136847609
|
1.526
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_+_86934525
|
1.511
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr13_+_24144402
|
1.507
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr9_+_103340335
|
1.494
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr3_+_151531860
|
1.493
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr20_+_11898536
|
1.482
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chrX_+_36065052
|
1.478
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr6_-_87804773
|
1.472
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr8_+_98703361
|
1.468
|
|
MTDH
|
metadherin
|
|
chr10_+_57358749
|
1.443
|
NM_001190478
|
MTRNR2L5
|
MT-RNR2-like 5
|
|
chr22_+_29449877
|
1.428
|
|
ZNRF3
|
zinc and ring finger 3
|
|
chr12_+_111843743
|
1.418
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr17_-_37350051
|
1.415
|
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr2_+_219472487
|
1.410
|
NM_032726
|
PLCD4
|
phospholipase C, delta 4
|
|
chrX_-_19688991
|
1.401
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_-_50595303
|
1.399
|
NM_001243775
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_+_162602308
|
1.377
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr15_+_25281105
|
1.375
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr10_-_14816895
|
1.364
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr12_+_21525801
|
1.351
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr2_-_231989755
|
1.346
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr2_+_85766286
|
1.342
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr5_-_64777703
|
1.341
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr8_+_55528626
|
1.335
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr5_+_131409484
|
1.320
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr11_+_33363104
|
1.317
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr2_-_224467142
|
1.313
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr1_+_197886499
|
1.298
|
NM_020204
|
LHX9
|
LIM homeobox 9
|
|
chr5_+_92918924
|
1.289
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr11_-_128457452
|
1.281
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_229569839
|
1.276
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr6_+_127898318
|
1.269
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr13_+_111855406
|
1.237
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr6_-_47009992
|
1.235
|
NM_025048
NM_153840
|
GPR110
|
G protein-coupled receptor 110
|
|
chr12_-_28122884
|
1.233
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr2_-_55237255
|
1.231
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr1_+_152943127
|
1.216
|
NM_173080
|
SPRR4
|
small proline-rich protein 4
|
|
chr5_+_180794268
|
1.212
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr4_-_152149042
|
1.211
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr6_-_123957941
|
1.209
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr8_-_95907423
|
1.206
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr3_-_197300193
|
1.202
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr1_+_40735749
|
1.202
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr2_+_170662044
|
1.198
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr12_-_53189780
|
1.191
|
NM_057088
|
KRT3
|
keratin 3
|
|
chr5_+_65372744
|
1.185
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr11_-_17191287
|
1.184
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr1_-_86043932
|
1.177
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_103378489
|
1.176
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr10_-_69425356
|
1.176
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr6_-_136847078
|
1.174
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr9_+_33026482
|
1.173
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr6_+_4890225
|
1.173
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chrX_+_17653412
|
1.159
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr11_+_57529233
|
1.151
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr5_-_88119604
|
1.150
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_-_18067445
|
1.148
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr20_+_58713521
|
1.145
|
|
LOC284757
|
uncharacterized LOC284757
|
|
chr1_-_161193375
|
1.144
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr4_-_71532206
|
1.135
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr1_-_183274008
|
1.132
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr2_-_225370687
|
1.130
|
|
CUL3
|
cullin 3
|
|
chr7_+_116660291
|
1.125
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr16_-_12010518
|
1.123
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr12_-_71031174
|
1.119
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chrX_-_133792497
|
1.118
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr11_-_7818488
|
1.116
|
NM_153444
|
OR5P2
|
olfactory receptor, family 5, subfamily P, member 2
|
|
chr7_-_50628744
|
1.113
|
NM_000790
NM_001242886
NM_001242887
NM_001242888
NM_001242889
NM_001242890
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr3_-_111314181
|
1.108
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr12_+_28410132
|
1.103
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr13_-_99959649
|
1.101
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr2_+_183608329
|
1.091
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chrX_+_37639246
|
1.089
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr8_-_101936521
|
1.075
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr4_-_80994309
|
1.061
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr18_-_51690988
|
1.060
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chrX_-_19817907
|
1.056
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_-_110723127
|
1.047
|
|
CFI
|
complement factor I
|
|
chr17_+_3379295
|
1.046
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr2_-_106055229
|
1.041
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr13_-_24007822
|
1.030
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chrX_+_41583407
|
1.026
|
NM_080817
|
GPR82
|
G protein-coupled receptor 82
|
|
chr1_+_207070787
|
1.022
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr13_-_75901897
|
1.019
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr15_-_55562483
|
1.018
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr7_+_28338939
|
1.014
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_-_224467095
|
1.006
|
|
SCG2
|
secretogranin II
|
|
chr2_+_102508333
|
1.005
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr15_+_29033259
|
1.004
|
|
LOC100289656
|
Dexi homolog (mouse) pseudogene
|
|
chr1_+_24834083
|
0.991
|
NM_001251978
|
RCAN3
|
RCAN family member 3
|
|
chr18_+_61582744
|
0.980
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr8_-_110342107
|
0.979
|
NM_001128211
|
NUDCD1
|
NudC domain containing 1
|
|
chr7_+_107110501
|
0.976
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chrX_+_49832214
|
0.973
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_-_93257950
|
0.970
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr17_-_29624249
|
0.968
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr10_+_69869249
|
0.967
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr13_+_76334796
|
0.966
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr4_-_146095891
|
0.961
|
NM_017493
|
OTUD4
|
OTU domain containing 4
|
|
chr6_+_142510627
|
0.959
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr6_+_151561094
|
0.956
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_131148544
|
0.954
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|
|
chr2_+_228356287
|
0.953
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr4_-_44653657
|
0.951
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr1_+_209859524
|
0.946
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr4_-_184241926
|
0.944
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr3_+_132316080
|
0.943
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr7_-_112727778
|
0.936
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_-_32403310
|
0.936
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr17_+_21308447
|
0.935
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr12_-_81763127
|
0.931
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_-_69536314
|
0.928
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr5_-_83016850
|
0.927
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr13_-_38172862
|
0.927
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr7_-_115608366
|
0.927
|
NM_001244583
|
TFEC
|
transcription factor EC
|
|
chr1_-_230991781
|
0.917
|
NM_001136495
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr16_-_67977956
|
0.913
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr11_-_60623422
|
0.906
|
NM_004778
|
PTGDR2
|
prostaglandin D2 receptor 2
|
|
chr5_-_32388595
|
0.900
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr5_+_94727047
|
0.898
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr8_-_8239188
|
0.897
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr11_+_113845796
|
0.897
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr1_-_207095198
|
0.895
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr2_-_37076016
|
0.891
|
|
STRN
|
striatin, calmodulin binding protein
|
|
chr5_+_68800003
|
0.891
|
NM_001205255
|
OCLN
|
occludin
|
|
chr12_-_52779362
|
0.886
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr3_+_132318980
|
0.882
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr2_-_43453737
|
0.882
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr12_-_123834984
|
0.876
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr8_-_102803196
|
0.872
|
|
NCALD
|
neurocalcin delta
|
|
chr15_-_99057610
|
0.861
|
NM_182562
|
FAM169B
|
family with sequence similarity 169, member B
|
|
chr2_-_163099877
|
0.860
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_-_98262217
|
0.859
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr10_-_69455926
|
0.856
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr13_-_46756326
|
0.844
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_-_21663908
|
0.843
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr4_+_77953942
|
0.840
|
|
SEPT11
|
septin 11
|
|
chr16_+_57050985
|
0.840
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr14_+_90863365
|
0.839
|
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr14_+_67831484
|
0.839
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr1_-_155948934
|
0.831
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr8_+_49984902
|
0.830
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|