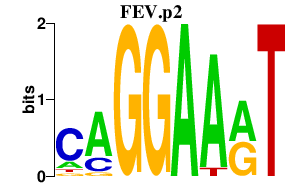
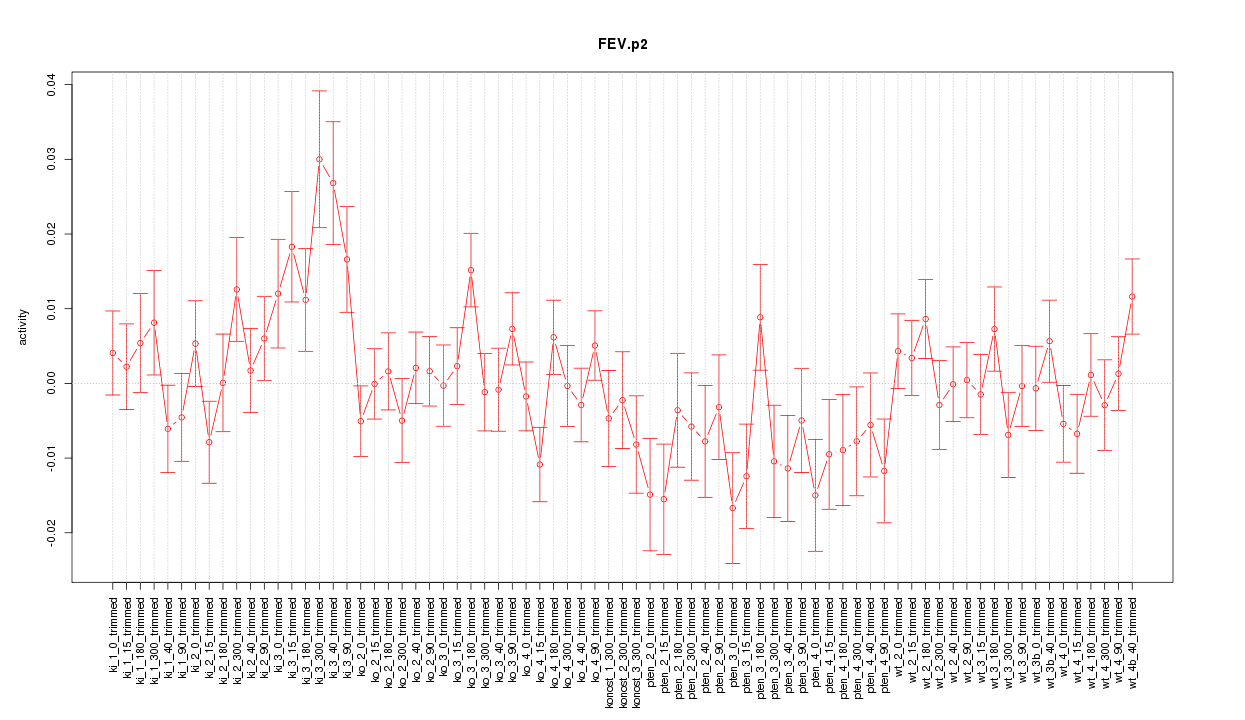
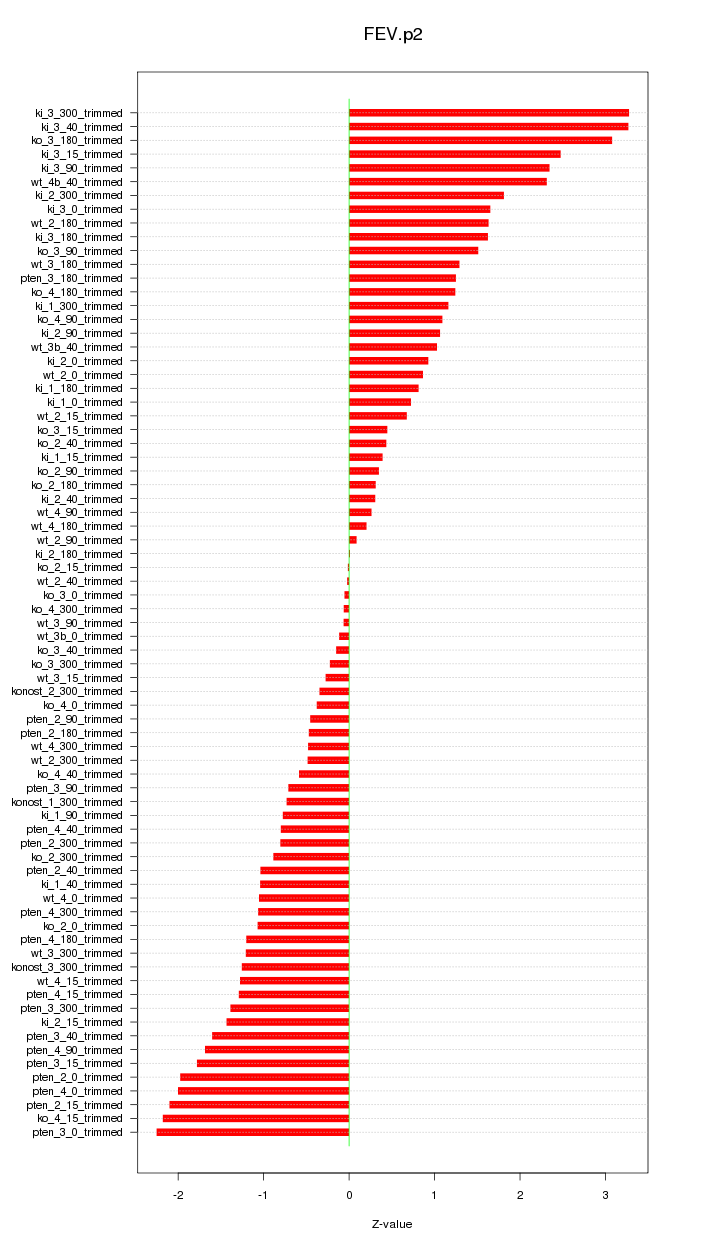
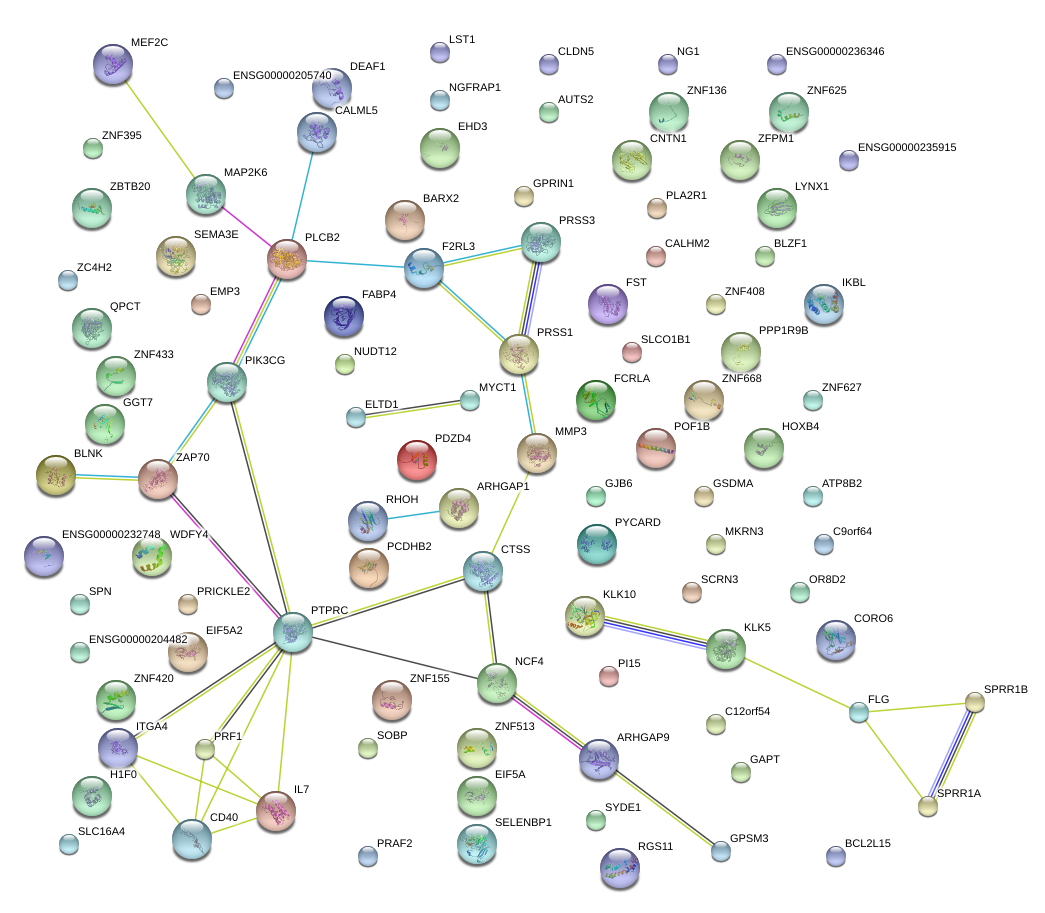
|
chr19_+_15218141
|
17.305
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr8_-_82395401
|
12.689
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_+_154300275
|
12.546
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr16_-_31214054
|
11.274
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chr20_+_44746889
|
9.323
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr17_+_67410825
|
9.231
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr20_+_44746948
|
8.715
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr9_-_86571533
|
7.911
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr5_+_140474198
|
7.662
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr2_+_37571752
|
7.466
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr5_-_102898450
|
6.986
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr1_-_151345156
|
6.516
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr19_-_51523272
|
6.298
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr19_-_51522953
|
5.990
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr17_-_46655544
|
5.944
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr7_+_106505739
|
5.914
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr20_-_33460558
|
5.855
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr19_-_51456159
|
5.854
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr2_+_182321933
|
5.779
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr7_+_106505696
|
5.744
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chrX_-_64196268
|
5.587
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr19_+_44488331
|
5.242
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chr3_-_64211111
|
5.187
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr12_-_123214882
|
5.184
|
NM_032554
|
HCAR1
|
hydroxycarboxylic acid receptor 1
|
|
chrX_+_102632108
|
5.157
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr15_-_40600025
|
5.112
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_-_110933635
|
5.106
|
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr8_-_79717540
|
5.030
|
NM_000880
NM_001199886
NM_001199887
NM_001199888
|
IL7
|
interleukin 7
|
|
chr6_-_32160637
|
4.981
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr11_-_46722115
|
4.967
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr7_+_106505876
|
4.931
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr2_+_175260455
|
4.860
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr17_+_38119225
|
4.607
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr12_-_57871631
|
4.583
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr15_-_40600118
|
4.568
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chrX_-_153072265
|
4.504
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr22_+_37257004
|
4.474
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr13_-_20805371
|
4.376
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_+_152956563
|
4.355
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr1_-_110933683
|
4.346
|
NM_001201546
NM_001201547
NM_001201548
NM_001201549
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr11_+_129245834
|
4.291
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr15_-_40599872
|
4.263
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_-_150738255
|
4.227
|
|
CTSS
|
cathepsin S
|
|
chr19_-_12267528
|
4.203
|
NM_145233
|
ZNF625-ZNF20
ZNF625
|
ZNF625-ZNF20 readthrough
zinc finger protein 625
|
|
chr19_+_48828820
|
4.136
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr6_-_32160289
|
4.118
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr2_+_175260505
|
4.093
|
|
SCRN3
|
secernin 3
|
|
chr12_+_48876285
|
4.065
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr3_-_170626381
|
4.048
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr19_+_48828627
|
3.952
|
NM_001425
|
EMP3
|
epithelial membrane protein 3
|
|
chr22_+_38201300
|
3.947
|
|
H1F0
|
H1 histone family, member 0
|
|
chr3_-_114343052
|
3.944
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_170626425
|
3.939
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr5_+_52776479
|
3.912
|
|
FST
|
follistatin
|
|
chr1_+_161676761
|
3.872
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr9_+_33750463
|
3.752
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chrX_-_48931645
|
3.749
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr1_-_114429996
|
3.724
|
|
BCL2L15
|
BCL2-like 15
|
|
chr19_+_37569307
|
3.654
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|
|
chr10_+_49892860
|
3.645
|
|
WDFY4
|
WDFY family member 4
|
|
chr19_+_16999871
|
3.594
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr16_-_31085503
|
3.569
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chrX_-_84634396
|
3.530
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr10_-_72362441
|
3.525
|
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr10_-_1034280
|
3.507
|
|
LOC100288619
|
uncharacterized LOC100288619
|
|
chr12_+_89744714
|
3.480
|
|
LOC100131490
|
uncharacterized LOC100131490
|
|
chr10_-_72362513
|
3.478
|
NM_001083116
NM_005041
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr11_-_46722160
|
3.478
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr5_-_176037091
|
3.464
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr2_+_182321618
|
3.456
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr8_+_75736771
|
3.338
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr10_-_105212149
|
3.315
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr4_+_40198526
|
3.314
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr15_+_23810808
|
3.287
|
|
MKRN3
|
makorin ring finger protein 3
|
|
chr6_+_153019029
|
3.268
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr8_-_143859638
|
3.267
|
NM_023946
NM_177457
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr11_+_46722564
|
3.236
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr6_+_31553955
|
3.189
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr16_+_29674564
|
3.187
|
|
SPN
|
sialophorin
|
|
chr1_+_198608216
|
3.186
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr5_+_57787260
|
3.162
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr16_+_29674568
|
3.148
|
NM_003123
|
SPN
|
sialophorin
|
|
chr11_-_102714240
|
3.127
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr2_-_160919120
|
3.126
|
NM_001007267
NM_001195641
NM_007366
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr10_-_98031154
|
3.120
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr5_-_88178964
|
3.090
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_+_70231163
|
3.086
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr16_+_29674282
|
3.073
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr17_-_27948345
|
3.068
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr1_+_169337193
|
3.034
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr7_-_83278478
|
3.033
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr1_-_79472360
|
2.994
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr6_+_107811272
|
2.991
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr2_+_31456981
|
2.959
|
|
EHD3
|
EH-domain containing 3
|
|
chr2_-_27603584
|
2.938
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr16_-_325867
|
2.916
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr10_-_5541401
|
2.914
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr2_+_98330030
|
2.892
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr12_-_57873628
|
2.871
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr22_-_19512859
|
2.857
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr12_+_41086189
|
2.836
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr14_+_77227783
|
2.831
|
|
|
|
|
chr1_+_169337432
|
2.823
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr19_-_58238895
|
2.810
|
NM_024833
|
ZNF671
|
zinc finger protein 671
|
|
chr11_-_65667860
|
2.810
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr11_-_64764388
|
2.800
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chrX_+_84498993
|
2.788
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr11_+_107799134
|
2.775
|
NM_017516
|
RAB39A
|
RAB39A, member RAS oncogene family
|
|
chr16_-_31214003
|
2.770
|
|
PYCARD
|
PYD and CARD domain containing
|
|
chr12_-_57873364
|
2.761
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr1_+_198608244
|
2.759
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_-_46722139
|
2.751
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr7_-_3083346
|
2.747
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr5_+_52776408
|
2.708
|
|
FST
|
follistatin
|
|
chr3_+_38080695
|
2.688
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr9_+_33795558
|
2.672
|
NM_002771
|
PRSS3
|
protease, serine, 3
|
|
chr5_+_178450775
|
2.669
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr19_-_17375522
|
2.669
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chrX_-_84634664
|
2.659
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr11_+_22688156
|
2.656
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr11_-_123848397
|
2.650
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr20_+_49126920
|
2.646
|
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|
|
chr5_-_54281345
|
2.639
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr5_+_180581942
|
2.639
|
NM_206880
|
OR2V2
|
olfactory receptor, family 2, subfamily V, member 2
|
|
chr20_+_45523247
|
2.637
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr1_-_202129705
|
2.633
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr1_-_39407449
|
2.633
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr1_+_198608136
|
2.629
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr3_+_113251217
|
2.629
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr20_-_56286540
|
2.616
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr10_+_81892429
|
2.608
|
|
PLAC9
|
placenta-specific 9
|
|
chr11_-_111412422
|
2.589
|
|
|
|
|
chr10_+_81892497
|
2.587
|
|
PLAC9
|
placenta-specific 9
|
|
chr3_-_183273407
|
2.581
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr2_+_31456879
|
2.574
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr17_+_4981721
|
2.573
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr17_+_4981754
|
2.573
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr19_-_36246411
|
2.571
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr4_+_152198324
|
2.569
|
NM_183375
|
PRSS48
|
protease, serine, 48
|
|
chr1_-_150738232
|
2.546
|
|
CTSS
|
cathepsin S
|
|
chr16_-_50715116
|
2.544
|
|
SNX20
|
sorting nexin 20
|
|
chr3_+_173116243
|
2.541
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr16_+_30483959
|
2.531
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr11_-_75141210
|
2.530
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr18_-_61329113
|
2.528
|
NM_006919
|
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
|
|
chrX_-_84634742
|
2.522
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chrX_-_64254592
|
2.522
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr19_+_16999807
|
2.522
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr17_-_16875388
|
2.512
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr20_+_49126908
|
2.509
|
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|
|
chr19_-_51413993
|
2.494
|
NM_004917
|
KLK4
|
kallikrein-related peptidase 4
|
|
chr16_+_30484049
|
2.491
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr1_-_150738292
|
2.489
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr19_+_12273866
|
2.488
|
NM_003437
|
ZNF136
|
zinc finger protein 136
|
|
chr16_+_1583666
|
2.487
|
|
TMEM204
|
transmembrane protein 204
|
|
chr20_+_43104513
|
2.487
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr11_-_31825593
|
2.486
|
|
PAX6
|
paired box 6
|
|
chr16_-_29757264
|
2.481
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr6_-_154677868
|
2.470
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr2_+_69001932
|
2.468
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr13_+_46039024
|
2.466
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr15_-_40599974
|
2.462
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr15_+_68924326
|
2.456
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr6_-_22297729
|
2.455
|
NM_000948
|
PRL
|
prolactin
|
|
chr14_+_63671101
|
2.445
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr12_-_6798619
|
2.444
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr9_-_21368005
|
2.441
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chrX_-_50557019
|
2.436
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr5_-_141993691
|
2.432
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_+_40506449
|
2.429
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr7_+_29234325
|
2.429
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr1_-_156698188
|
2.428
|
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2
|
|
chr20_+_49126919
|
2.410
|
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|
|
chr17_-_42466840
|
2.382
|
NM_000419
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41)
|
|
chr7_+_100770378
|
2.379
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr6_-_11779279
|
2.363
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr2_-_27603249
|
2.362
|
NM_001201459
|
ZNF513
|
zinc finger protein 513
|
|
chrX_-_70331297
|
2.359
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr11_-_102651321
|
2.353
|
NM_002425
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr7_-_45018559
|
2.351
|
|
MYO1G
|
myosin IG
|
|
chr2_-_238322770
|
2.346
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrY_+_2709630
|
2.345
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr15_-_40600065
|
2.344
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr3_+_111260925
|
2.339
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr12_-_6798528
|
2.327
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr1_+_209859524
|
2.326
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chrY_+_2709596
|
2.326
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr11_-_125773086
|
2.322
|
NM_031307
|
PUS3
|
pseudouridylate synthase 3
|
|
chr15_-_41120905
|
2.317
|
NM_001130143
NM_017726
|
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D
|
|
chr9_+_34458810
|
2.313
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr5_+_140625146
|
2.309
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chr3_-_50649080
|
2.302
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr19_-_36246206
|
2.299
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr3_+_11267666
|
2.297
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr11_+_67171379
|
2.296
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr10_-_28287790
|
2.292
|
NM_018076
|
ARMC4
|
armadillo repeat containing 4
|
|
chr17_-_9939873
|
2.289
|
|
GAS7
|
growth arrest-specific 7
|
|
chr22_-_17700265
|
2.288
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr14_+_21249209
|
2.285
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr1_+_40506406
|
2.278
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|