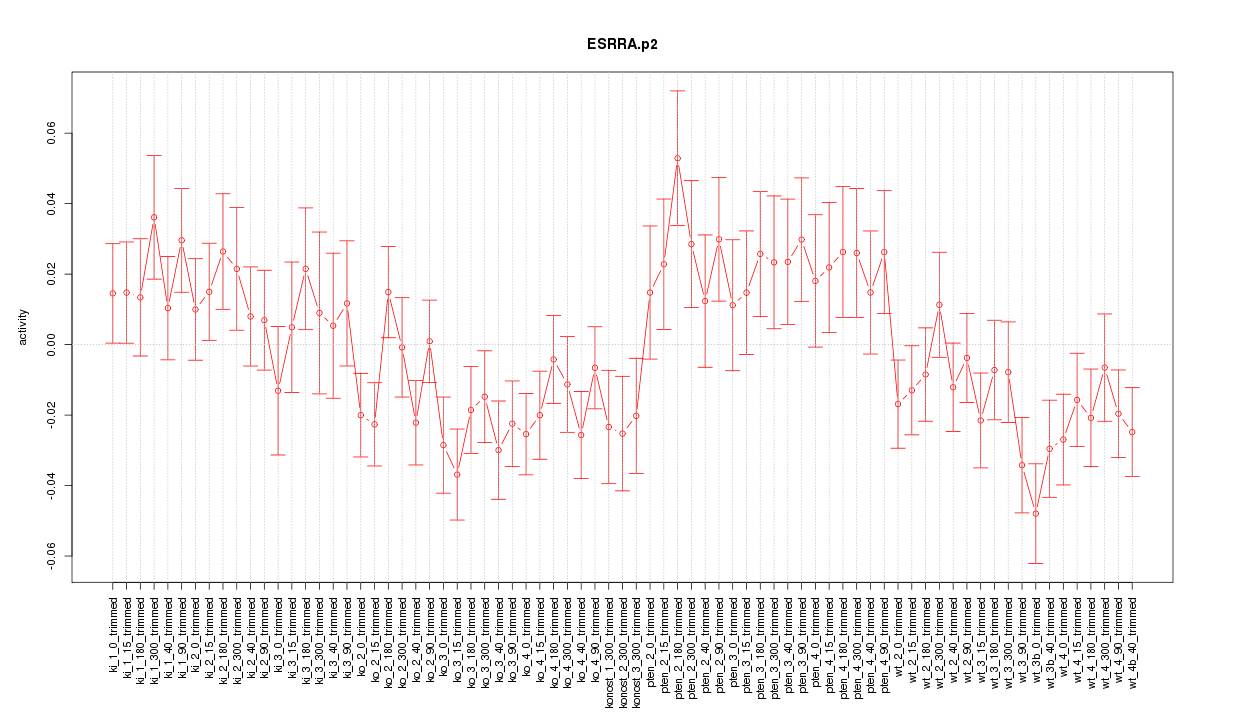
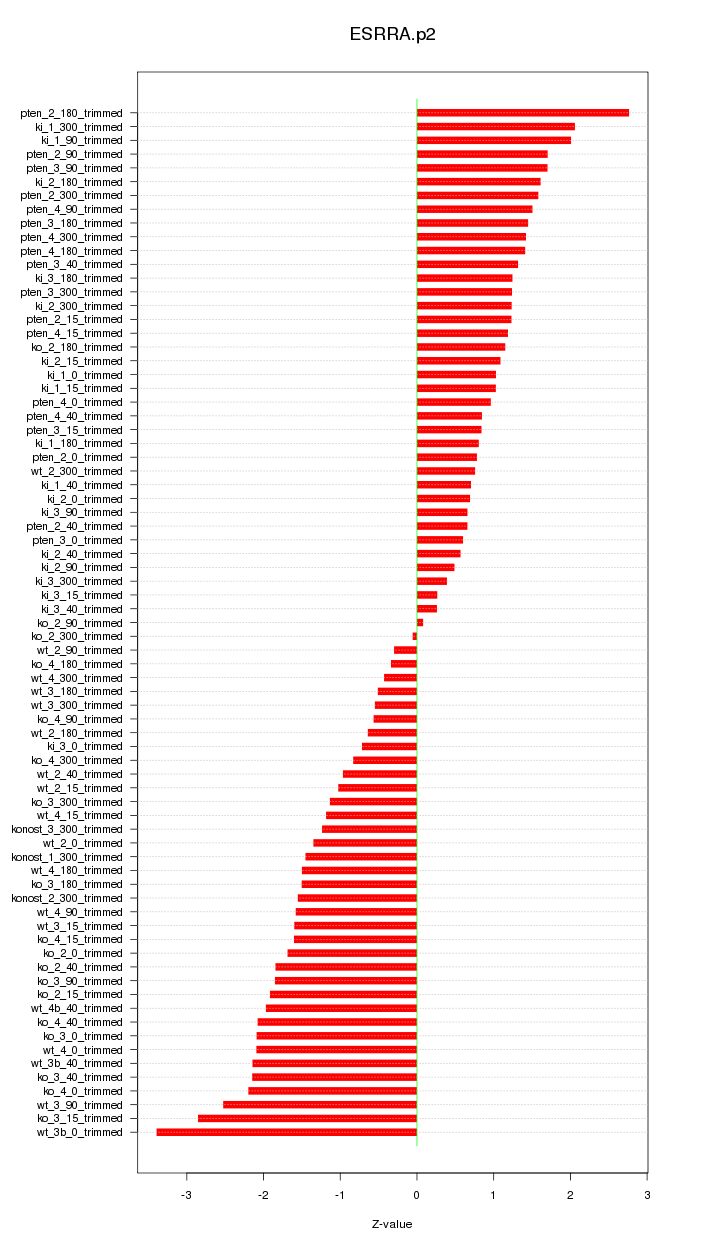
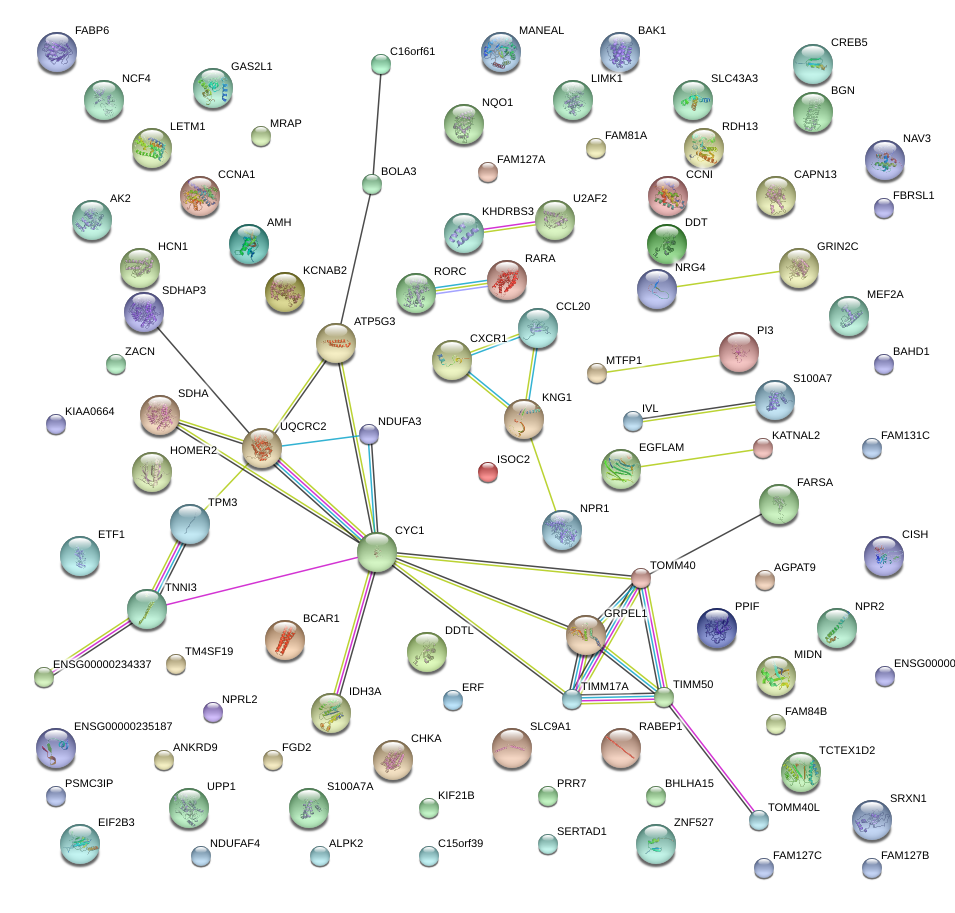
|
chr19_+_1776117
|
13.131
|
|
|
|
|
chr15_-_76352068
|
11.809
|
|
NRG4
|
neuregulin 4
|
|
chr2_-_31030271
|
8.268
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr19_+_2249112
|
7.692
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr19_-_55574434
|
7.161
|
NM_001145971
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr10_+_81107242
|
6.639
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chrX_+_49047887
|
5.724
|
|
|
|
|
chr10_+_81107259
|
5.663
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr2_+_228678556
|
5.625
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr3_-_50649080
|
5.500
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr1_+_152881022
|
5.397
|
NM_005547
|
IVL
|
involucrin
|
|
chr5_+_176875325
|
5.226
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr7_+_97841565
|
5.208
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr18_+_44497512
|
5.132
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr22_+_37257004
|
4.819
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr1_-_153433104
|
4.262
|
NM_002963
|
S100A7
|
S100 calcium binding protein A7
|
|
chr19_+_45394642
|
4.228
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr15_+_59730272
|
3.966
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr19_+_54606141
|
3.937
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr17_+_38474472
|
3.915
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr5_+_159656436
|
3.879
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr6_-_33547833
|
3.673
|
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr19_-_40931880
|
3.616
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr10_+_81107219
|
3.602
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_+_81107228
|
3.567
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr19_-_36019194
|
3.551
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr20_-_633808
|
3.470
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr2_-_176046120
|
3.452
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr1_+_153388999
|
3.363
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr1_-_200992824
|
3.349
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr5_+_38445577
|
3.283
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr17_-_72855884
|
3.272
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr4_+_84457390
|
3.144
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr3_+_186435097
|
3.130
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr1_+_38259458
|
3.116
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr19_+_41116069
|
3.068
|
|
|
|
|
chr8_+_136469557
|
3.042
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr20_-_633844
|
3.028
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr12_+_78225068
|
2.947
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr22_-_43036606
|
2.944
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr19_-_55973048
|
2.924
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr15_+_75494235
|
2.908
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr15_+_75494233
|
2.902
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr19_+_37862055
|
2.886
|
NM_032453
|
ZNF527
|
zinc finger protein 527
|
|
chr19_+_45394522
|
2.850
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr6_-_97345747
|
2.837
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr14_-_102975996
|
2.804
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr15_+_75494196
|
2.803
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr2_-_74374998
|
2.795
|
NM_001035505
NM_212552
|
BOLA3
|
bolA homolog 3 (E. coli)
|
|
chr1_-_27481400
|
2.758
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr22_+_24373107
|
2.742
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chr6_+_36973409
|
2.730
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr6_-_97345676
|
2.698
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chrX_-_134156488
|
2.696
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr7_+_28530747
|
2.661
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr19_-_55668956
|
2.640
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr7_+_48128738
|
2.607
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr13_+_37006628
|
2.605
|
|
CCNA1
|
cyclin A1
|
|
chr17_-_2614926
|
2.601
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr11_-_67888857
|
2.596
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr19_-_55972997
|
2.593
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr12_+_133067156
|
2.526
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr8_+_145149985
|
2.503
|
|
CYC1
|
cytochrome c-1
|
|
chr5_+_218441
|
2.448
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr4_-_7069755
|
2.447
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr7_+_73498318
|
2.441
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr2_-_219031646
|
2.437
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr19_+_39971496
|
2.412
|
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr19_+_1248548
|
2.404
|
NM_177401
|
MIDN
|
midnolin
|
|
chrX_+_152760346
|
2.401
|
NM_001711
|
BGN
|
biglycan
|
|
chr1_-_45452215
|
2.399
|
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr22_+_29702950
|
2.397
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr15_+_78441737
|
2.381
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr1_-_33502484
|
2.321
|
NM_001199199
NM_001625
NM_013411
|
AK2
|
adenylate kinase 2
|
|
chr5_+_218355
|
2.318
|
NM_004168
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr1_+_6094342
|
2.293
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr5_-_45696219
|
2.289
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr19_-_13044459
|
2.286
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr16_-_75285383
|
2.250
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr5_+_218447
|
2.243
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr1_-_151798552
|
2.237
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr9_+_35792405
|
2.217
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr16_-_69760444
|
2.216
|
NM_000903
NM_001025433
NM_001025434
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr6_-_97345760
|
2.205
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr1_-_16399936
|
2.161
|
NM_182623
|
FAM131C
|
family with sequence similarity 131, member C
|
|
chr11_-_67888670
|
2.155
|
|
CHKA
|
choline kinase alpha
|
|
chr22_+_30821610
|
2.144
|
NM_001003704
NM_016498
|
MTFP1
|
mitochondrial fission process 1
|
|
chr15_-_83621350
|
2.139
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr19_-_55972935
|
2.138
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr15_+_100173182
|
2.137
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr5_+_218428
|
2.121
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr16_-_81040439
|
2.121
|
NM_020188
|
C16orf61
|
chromosome 16 open reading frame 61
|
|
chr11_-_60623422
|
2.090
|
NM_004778
|
PTGDR2
|
prostaglandin D2 receptor 2
|
|
chr14_-_102976096
|
2.068
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr20_+_43803539
|
2.066
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr1_-_33502470
|
2.057
|
|
AK2
|
adenylate kinase 2
|
|
chr1_-_154164551
|
2.048
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr19_-_42759151
|
2.031
|
|
ERF
|
Ets2 repressor factor
|
|
chr1_+_161195832
|
2.029
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr8_-_127570465
|
2.028
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr16_-_69760354
|
1.980
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr3_-_196065191
|
1.973
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr16_-_69760367
|
1.972
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr19_+_56165415
|
1.971
|
NM_001012478
NM_007279
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chrX_+_152760452
|
1.963
|
|
BGN
|
biglycan
|
|
chr4_-_1857677
|
1.963
|
NM_012318
|
LETM1
|
leucine zipper-EF-hand containing transmembrane protein 1
|
|
chr11_-_57194591
|
1.961
|
NM_199329
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr1_+_201924618
|
1.942
|
NM_006335
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast)
|
|
chr22_+_24309025
|
1.940
|
NM_001084393
|
DDTL
|
D-dopachrome tautomerase-like
|
|
chr17_+_74075262
|
1.931
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr15_+_78441733
|
1.918
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr17_-_40729649
|
1.915
|
NM_013290
NM_016556
|
PSMC3IP
|
PSMC3 interacting protein
|
|
chr18_-_56296188
|
1.914
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr16_+_21964700
|
1.909
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr15_+_40733302
|
1.907
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr17_+_5185551
|
1.906
|
NM_001083585
NM_004703
|
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1
|
|
chr11_-_34379526
|
1.894
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr15_+_43885251
|
1.886
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr22_+_31518923
|
1.884
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr16_+_21964608
|
1.880
|
NM_003366
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr1_-_33502452
|
1.867
|
|
AK2
|
adenylate kinase 2
|
|
chr8_-_127570710
|
1.855
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr19_+_1383898
|
1.855
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr1_-_33502428
|
1.853
|
|
AK2
|
adenylate kinase 2
|
|
chr19_+_39616384
|
1.852
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr2_-_106055229
|
1.840
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr19_+_1383877
|
1.834
|
NM_024407
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr11_-_57194556
|
1.831
|
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr11_-_57194585
|
1.825
|
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr10_-_76995685
|
1.822
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr1_+_32757711
|
1.811
|
|
HDAC1
|
histone deacetylase 1
|
|
chr3_-_113465101
|
1.805
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr19_+_1383904
|
1.797
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr19_-_474875
|
1.789
|
NM_182577
|
ODF3L2
|
outer dense fiber of sperm tails 3-like 2
|
|
chr22_-_25758523
|
1.788
|
NM_182492
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like
|
|
chr15_+_43985083
|
1.786
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr9_+_37650996
|
1.786
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr3_+_184038050
|
1.777
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr16_+_21964677
|
1.777
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr18_-_5543967
|
1.767
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr10_+_99185977
|
1.750
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr3_-_10067989
|
1.740
|
|
CIDECP
|
cell death-inducing DFFA-like effector c pseudogene
|
|
chr10_+_75757847
|
1.732
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr19_-_39826676
|
1.719
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr5_+_34684611
|
1.711
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr15_-_76304762
|
1.709
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr15_+_74466887
|
1.703
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr11_-_57298180
|
1.700
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr19_+_46850344
|
1.696
|
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr16_+_57050985
|
1.694
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr16_-_85832900
|
1.690
|
NM_001142288
NM_006067
|
COX4NB
|
COX4 neighbor
|
|
chr11_+_18194383
|
1.689
|
NM_054032
|
MRGPRX4
|
MAS-related GPR, member X4
|
|
chr22_+_35779103
|
1.683
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr12_+_119616594
|
1.682
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr7_+_94536926
|
1.680
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr8_+_104831415
|
1.674
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr22_+_32010152
|
1.669
|
|
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast)
|
|
chr15_+_78441678
|
1.665
|
NM_005530
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr11_-_67442018
|
1.663
|
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chrX_-_129299629
|
1.658
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr7_+_48128939
|
1.649
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr11_-_111957417
|
1.634
|
NM_012459
|
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast)
|
|
chr1_-_33502464
|
1.634
|
|
AK2
|
adenylate kinase 2
|
|
chr9_-_130889993
|
1.627
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr20_+_30865435
|
1.619
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr20_-_3748355
|
1.609
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr10_+_99186040
|
1.606
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr1_+_32757702
|
1.589
|
|
HDAC1
|
histone deacetylase 1
|
|
chr19_-_1513187
|
1.582
|
NM_213604
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr19_+_46850245
|
1.581
|
NM_001204284
NM_006247
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr2_+_234621637
|
1.580
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chrX_-_129299680
|
1.579
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr2_+_202125222
|
1.575
|
NM_033356
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr10_+_99186004
|
1.572
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr1_+_40997232
|
1.571
|
NM_152373
|
ZNF684
|
zinc finger protein 684
|
|
chr8_+_145149978
|
1.569
|
|
CYC1
|
cytochrome c-1
|
|
chr11_-_65793849
|
1.567
|
NM_053054
|
CATSPER1
|
cation channel, sperm associated 1
|
|
chr10_+_105156430
|
1.565
|
|
PDCD11
|
programmed cell death 11
|
|
chr7_+_129008044
|
1.563
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chrX_-_129299803
|
1.562
|
NM_001130847
NM_004208
NM_145812
NM_145813
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr16_+_89989686
|
1.561
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr10_+_99186017
|
1.556
|
NM_002629
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr20_-_13765509
|
1.556
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr3_-_197354629
|
1.548
|
|
LOC220729
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) pseudogene
|
|
chr11_-_57194512
|
1.541
|
NM_014096
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr10_+_99186012
|
1.539
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr8_+_145149936
|
1.530
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr19_-_39322411
|
1.523
|
NM_001398
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal
|
|
chr11_+_118272103
|
1.519
|
NM_006476
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G
|
|
chr11_-_67442086
|
1.515
|
NM_000695
|
ALDH3B2
|
aldehyde dehydrogenase 3 family, member B2
|
|
chr18_-_44497307
|
1.515
|
NM_004671
NM_173206
|
PIAS2
|
protein inhibitor of activated STAT, 2
|
|
chr10_+_99186042
|
1.509
|
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr19_+_3185860
|
1.489
|
NM_020170
|
NCLN
|
nicalin
|
|
chr16_+_691827
|
1.487
|
NM_138418
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr16_+_718080
|
1.481
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr18_-_43678233
|
1.472
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr19_-_1512997
|
1.469
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr14_+_100150596
|
1.467
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr4_+_2470846
|
1.464
|
|
RNF4
|
ring finger protein 4
|
|
chr3_-_27763784
|
1.456
|
NM_005442
|
EOMES
|
eomesodermin
|