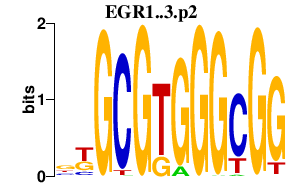
|
chr20_-_62462568
|
12.410
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr1_-_9189087
|
10.054
|
NM_024980
|
GPR157
|
G protein-coupled receptor 157
|
|
chr6_+_126070813
|
7.784
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr17_+_56065408
|
7.553
|
|
|
|
|
chr19_+_8429010
|
7.491
|
NM_001039667
NM_139314
|
ANGPTL4
|
angiopoietin-like 4
|
|
chr11_+_129245834
|
7.385
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr10_-_103535656
|
7.326
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr2_+_241375207
|
7.298
|
|
GPC1
|
glypican 1
|
|
chr12_-_28122884
|
6.992
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr8_-_143695812
|
6.580
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr19_-_1863425
|
6.395
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr3_-_39195101
|
5.941
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr19_+_1249882
|
5.854
|
|
MIDN
|
midnolin
|
|
chr12_+_57482886
|
5.799
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr2_-_46384
|
5.477
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr7_-_559030
|
5.464
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr3_-_39195034
|
5.419
|
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr6_+_37137882
|
5.409
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr6_+_126070895
|
5.361
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chrX_+_9432980
|
5.172
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr11_-_65667858
|
4.960
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_+_57482674
|
4.811
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr13_-_20767100
|
4.797
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr12_+_57482922
|
4.744
|
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr13_-_20767036
|
4.720
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr11_-_65667860
|
4.696
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_16435734
|
4.677
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr16_-_402617
|
4.674
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr19_-_51568323
|
4.610
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr1_+_6673744
|
4.592
|
NM_153812
|
PHF13
|
PHD finger protein 13
|
|
chr12_-_120806893
|
4.518
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr11_-_65667809
|
4.512
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr3_-_52090090
|
4.491
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr16_-_28074776
|
4.456
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr1_-_19282773
|
4.326
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr8_-_103666018
|
4.319
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr12_+_130822590
|
4.181
|
|
PIWIL1
|
piwi-like 1 (Drosophila)
|
|
chr2_+_105471745
|
4.151
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr3_+_11196213
|
4.124
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr6_+_126070738
|
4.070
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr10_-_108924287
|
3.965
|
NM_001013031
NM_001206569
NM_001206570
NM_001206571
NM_001206572
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr1_-_38471170
|
3.918
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr15_+_32322690
|
3.855
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr19_-_1567875
|
3.848
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr19_-_40931880
|
3.844
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr15_-_76352068
|
3.837
|
|
NRG4
|
neuregulin 4
|
|
chr10_+_5566923
|
3.822
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr7_-_106301329
|
3.748
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr19_+_16435632
|
3.744
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr1_-_99470177
|
3.729
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr7_-_44365019
|
3.704
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr1_+_15250578
|
3.685
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr19_-_10047014
|
3.672
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr4_+_1005609
|
3.560
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr17_-_43394326
|
3.548
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chrX_-_17879355
|
3.524
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr14_-_38064440
|
3.504
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr19_-_1237840
|
3.462
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr17_+_81037504
|
3.433
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chrX_-_152939256
|
3.410
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr22_+_39853316
|
3.408
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr8_-_22550057
|
3.392
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr1_-_223537400
|
3.374
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr6_+_126070724
|
3.369
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr2_-_204399906
|
3.340
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr10_+_129705299
|
3.339
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr17_+_30813928
|
3.316
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr7_+_55955129
|
3.290
|
|
ZNF713
|
zinc finger protein 713
|
|
chr11_-_67141008
|
3.289
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr16_+_69139466
|
3.288
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr10_+_122216465
|
3.282
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr3_-_171177851
|
3.266
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr18_-_45935627
|
3.257
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr5_+_7396030
|
3.252
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr18_+_12947972
|
3.247
|
NM_001013437
NM_031216
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr9_+_17578952
|
3.227
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr11_-_64545933
|
3.215
|
NM_001178030
|
SF1
|
splicing factor 1
|
|
chr10_+_129705359
|
3.177
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr19_-_4065467
|
3.171
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr16_+_84853585
|
3.169
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr14_-_53417636
|
3.165
|
|
FERMT2
|
fermitin family member 2
|
|
chr16_+_1203240
|
3.164
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr15_-_74726000
|
3.151
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr20_+_49348050
|
3.142
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr22_+_39853534
|
3.085
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr18_+_8609670
|
3.085
|
|
|
|
|
chr7_-_82073020
|
3.073
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr18_+_8609407
|
3.055
|
NM_001025300
|
RAB12
|
RAB12, member RAS oncogene family
|
|
chr18_+_12948004
|
3.054
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr5_-_134914447
|
3.008
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr1_+_210406194
|
3.005
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr15_+_40733302
|
2.988
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr5_-_72743829
|
2.986
|
|
FOXD1
|
forkhead box D1
|
|
chr8_-_22549901
|
2.984
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr10_-_77161136
|
2.966
|
|
ZNF503
|
zinc finger protein 503
|
|
chr1_+_6484816
|
2.966
|
NM_031475
|
ESPN
|
espin
|
|
chr11_-_67141647
|
2.956
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr11_-_64545988
|
2.949
|
|
SF1
|
splicing factor 1
|
|
chr10_+_134000335
|
2.938
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chrX_+_152770083
|
2.922
|
|
BGN
|
biglycan
|
|
chr7_-_559479
|
2.919
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr10_-_77159851
|
2.894
|
|
ZNF503
|
zinc finger protein 503
|
|
chr1_-_115212628
|
2.836
|
NM_198459
|
DENND2C
|
DENN/MADD domain containing 2C
|
|
chr4_-_819879
|
2.819
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr15_+_25093355
|
2.815
|
|
|
|
|
chr14_-_53417808
|
2.813
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr4_+_1005395
|
2.806
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr1_-_166135978
|
2.737
|
|
|
|
|
chr1_+_15736422
|
2.734
|
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr16_-_17564737
|
2.731
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr16_+_69141442
|
2.723
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr17_+_77751929
|
2.723
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr7_-_82072804
|
2.721
|
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr19_-_42759279
|
2.720
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr3_+_170075449
|
2.706
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr16_+_27325244
|
2.695
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr7_-_32931409
|
2.679
|
NM_015483
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr21_+_45875368
|
2.674
|
NM_030891
|
LRRC3
|
leucine rich repeat containing 3
|
|
chr12_-_76425207
|
2.672
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chrX_+_152760346
|
2.666
|
NM_001711
|
BGN
|
biglycan
|
|
chr15_-_83953465
|
2.664
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr4_+_7194317
|
2.599
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr7_-_32931363
|
2.596
|
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chrX_-_19905449
|
2.584
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr16_+_25703346
|
2.583
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr1_-_1475739
|
2.577
|
NM_001114748
|
TMEM240
|
transmembrane protein 240
|
|
chr12_-_109125254
|
2.565
|
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr17_-_78450247
|
2.564
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr12_+_130822432
|
2.563
|
NM_001190971
NM_004764
|
PIWIL1
|
piwi-like 1 (Drosophila)
|
|
chr5_-_72744351
|
2.558
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr15_+_31619043
|
2.554
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr11_-_64545864
|
2.530
|
|
SF1
|
splicing factor 1
|
|
chr16_+_27325269
|
2.527
|
|
IL4R
|
interleukin 4 receptor
|
|
chr17_+_38474513
|
2.500
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr3_-_45267804
|
2.482
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chrX_-_47479229
|
2.462
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr19_+_51815095
|
2.449
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr7_-_130418859
|
2.431
|
NM_138693
|
KLF14
|
Kruppel-like factor 14
|
|
chr5_-_176037091
|
2.425
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr3_-_50649080
|
2.408
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr9_-_138798957
|
2.406
|
NM_015447
|
CAMSAP1
|
calmodulin regulated spectrin-associated protein 1
|
|
chr17_+_38474472
|
2.405
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr1_-_38273823
|
2.384
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr11_-_70963619
|
2.374
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_+_1611544
|
2.369
|
|
FOXC1
|
forkhead box C1
|
|
chr22_+_41777932
|
2.369
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr8_-_8175860
|
2.360
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr17_+_38474544
|
2.358
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr22_+_38035694
|
2.346
|
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr19_-_4066739
|
2.341
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr12_+_111843743
|
2.337
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_+_39456887
|
2.313
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr4_-_6202251
|
2.306
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr18_+_31158540
|
2.305
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr16_-_402398
|
2.289
|
|
AXIN1
|
axin 1
|
|
chr3_-_50649110
|
2.275
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr14_-_38064176
|
2.275
|
|
FOXA1
|
forkhead box A1
|
|
chr17_+_30593194
|
2.269
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr19_-_1863546
|
2.268
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr19_-_42759151
|
2.261
|
|
ERF
|
Ets2 repressor factor
|
|
chr18_+_43913906
|
2.260
|
|
RNF165
|
ring finger protein 165
|
|
chr19_+_1753661
|
2.256
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr11_-_94964209
|
2.255
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr20_-_4229519
|
2.252
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr8_-_144241918
|
2.251
|
NM_001130478
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr12_-_96793797
|
2.241
|
|
CDK17
|
cyclin-dependent kinase 17
|
|
chr10_-_77160650
|
2.240
|
|
ZNF503
|
zinc finger protein 503
|
|
chr2_-_70781146
|
2.230
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr20_-_50159037
|
2.228
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr15_-_74726258
|
2.223
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr18_-_31158150
|
2.217
|
|
|
|
|
chr17_-_882905
|
2.209
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr15_+_73976537
|
2.205
|
|
|
|
|
chr4_-_38666429
|
2.202
|
|
FLJ13197
|
uncharacterized FLJ13197
|
|
chr5_-_176924601
|
2.201
|
NM_005451
NM_203352
NM_213636
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr18_+_12948010
|
2.196
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr14_-_71275732
|
2.192
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_-_38273846
|
2.181
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr14_-_38064299
|
2.167
|
|
FOXA1
|
forkhead box A1
|
|
chr6_+_30130982
|
2.142
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr12_-_51477138
|
2.140
|
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr19_+_1285766
|
2.136
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr7_+_45614179
|
2.135
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr17_-_79876027
|
2.129
|
|
SIRT7
|
sirtuin 7
|
|
chr12_-_76425375
|
2.127
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_+_119302709
|
2.121
|
|
|
|
|
chr11_+_107799134
|
2.115
|
NM_017516
|
RAB39A
|
RAB39A, member RAS oncogene family
|
|
chr20_+_20348744
|
2.114
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr8_-_143867931
|
2.112
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr8_-_141645603
|
2.104
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr11_+_119076974
|
2.100
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr6_+_83073501
|
2.099
|
|
TPBG
|
trophoblast glycoprotein
|
|
chr3_-_53079959
|
2.093
|
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr8_+_16884745
|
2.080
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr15_+_38544119
|
2.077
|
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr11_+_120382420
|
2.068
|
|
GRIK4
|
glutamate receptor, ionotropic, kainate 4
|
|
chr13_-_38443859
|
2.066
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr2_-_96810775
|
2.060
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr2_-_88752052
|
2.059
|
NM_001135649
|
FOXI3
|
forkhead box I3
|
|
chr16_+_84178864
|
2.057
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|