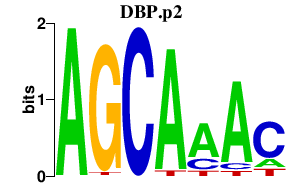
|
chr10_-_64576123
|
11.991
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr1_+_212788359
|
10.952
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_64576074
|
10.820
|
|
EGR2
|
early growth response 2
|
|
chr3_-_187452669
|
10.382
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr8_-_22550057
|
8.431
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr11_-_16424391
|
8.404
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_-_22550456
|
8.370
|
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
8.248
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_-_108231122
|
7.337
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr16_-_79634610
|
7.217
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr1_-_40367539
|
6.900
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr5_-_24645084
|
5.782
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr4_-_186732208
|
5.756
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_212738675
|
5.682
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr4_-_23891608
|
5.578
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr5_-_124080773
|
5.521
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr9_-_89562103
|
5.357
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr6_-_119470336
|
5.296
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr17_+_26850958
|
5.264
|
NM_003593
|
FOXN1
|
forkhead box N1
|
|
chr5_-_95768660
|
5.197
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr16_+_23194037
|
4.984
|
NM_001039
|
SCNN1G
|
sodium channel, nonvoltage-gated 1, gamma
|
|
chr4_-_186732047
|
4.816
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_136172802
|
4.678
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_145274916
|
4.638
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_-_90096308
|
4.636
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr14_-_73360792
|
4.482
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr16_+_30960434
|
4.286
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr16_+_86612114
|
4.224
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr5_+_145317297
|
4.203
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr16_+_30960413
|
4.132
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr6_-_134498973
|
4.089
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr16_+_30960377
|
4.044
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr9_-_20622433
|
3.860
|
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr11_+_113930430
|
3.732
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr2_+_166428845
|
3.576
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr1_-_150780705
|
3.440
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr3_-_52869234
|
3.426
|
NM_205853
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr12_-_88423145
|
3.419
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr20_+_51588945
|
3.388
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr4_-_159094163
|
3.379
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_+_54447660
|
3.312
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr2_-_192711651
|
3.287
|
|
SDPR
|
serum deprivation response
|
|
chr1_-_173176314
|
3.268
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_-_134495944
|
3.268
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_-_159093717
|
3.253
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr3_-_114343791
|
3.199
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_20622476
|
3.132
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr12_-_62586547
|
3.126
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr15_+_48009510
|
3.119
|
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr3_-_93747453
|
3.116
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr20_-_61002588
|
2.986
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr13_+_97874540
|
2.969
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_-_46142955
|
2.967
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr6_+_88054570
|
2.940
|
NM_001010868
|
C6orf163
|
chromosome 6 open reading frame 163
|
|
chr3_+_113616317
|
2.933
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr4_-_159093523
|
2.930
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_-_137071462
|
2.926
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr1_-_161039649
|
2.918
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_+_7017795
|
2.899
|
NM_080657
|
RSAD2
|
radical S-adenosyl methionine domain containing 2
|
|
chr20_+_43343523
|
2.898
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chrX_-_10851808
|
2.885
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr5_-_34043234
|
2.866
|
NM_030945
NM_181435
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3
|
|
chr13_+_97874704
|
2.848
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_+_89156804
|
2.827
|
|
EPHA3
|
EPH receptor A3
|
|
chr2_-_109605668
|
2.827
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr15_+_66995414
|
2.801
|
|
SMAD6
|
SMAD family member 6
|
|
chr1_-_151804225
|
2.784
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr1_-_92952429
|
2.766
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr6_-_149806025
|
2.748
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr14_-_23479328
|
2.737
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr3_-_49459870
|
2.671
|
|
AMT
|
aminomethyltransferase
|
|
chr6_+_126240369
|
2.664
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_-_190927321
|
2.629
|
NM_005259
|
MSTN
|
myostatin
|
|
chr7_-_121944485
|
2.622
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr3_-_12200850
|
2.610
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr3_-_143567163
|
2.543
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr12_+_56473944
|
2.540
|
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr1_+_205236591
|
2.537
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr7_-_27183225
|
2.523
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr1_+_164528914
|
2.506
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_+_20044362
|
2.491
|
|
NAV2
|
neuron navigator 2
|
|
chr3_+_111260925
|
2.479
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr3_+_69812620
|
2.472
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_72409191
|
2.470
|
NM_001172109
NM_001172110
|
SENP8
|
SUMO/sentrin specific peptidase family member 8
|
|
chr3_+_69812961
|
2.458
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_159092509
|
2.437
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_+_51435641
|
2.406
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr20_-_21378046
|
2.391
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr14_-_23652840
|
2.381
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr5_-_137071703
|
2.375
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr17_+_57915192
|
2.349
|
|
|
|
|
chr22_-_42828362
|
2.349
|
NM_145912
|
NFAM1
|
NFAT activating protein with ITAM motif 1
|
|
chr3_-_187463227
|
2.341
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_+_26225382
|
2.341
|
NM_003532
|
HIST1H3E
|
histone cluster 1, H3e
|
|
chr15_+_59904090
|
2.339
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr6_-_27858569
|
2.320
|
NM_003535
|
HIST1H3J
|
histone cluster 1, H3j
|
|
chr3_-_187463474
|
2.317
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr4_+_159442865
|
2.303
|
NM_001253727
NM_001253728
NM_001253729
NM_001253730
NM_001253732
NM_001253733
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr11_+_94706780
|
2.294
|
NM_018039
|
KDM4D
|
lysine (K)-specific demethylase 4D
|
|
chr6_-_42419782
|
2.280
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr13_-_48987017
|
2.279
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr12_-_9760481
|
2.274
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr17_+_28256873
|
2.268
|
NM_001145053
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr1_+_154540250
|
2.263
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr1_-_116383692
|
2.244
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr9_+_109625347
|
2.236
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr14_-_23652818
|
2.235
|
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr3_+_186330844
|
2.227
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr12_-_86229981
|
2.224
|
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr22_+_38201113
|
2.218
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr9_-_95298251
|
2.199
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr11_+_1718424
|
2.195
|
NM_001012416
|
KRTAP5-6
|
keratin associated protein 5-6
|
|
chr4_-_82393025
|
2.194
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chrX_-_135749436
|
2.190
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_+_150027837
|
2.188
|
|
SYNPO
|
synaptopodin
|
|
chr19_+_663381
|
2.180
|
|
|
|
|
chr1_-_221915393
|
2.171
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr7_+_22766765
|
2.167
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr5_-_160279045
|
2.160
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr10_-_104178900
|
2.156
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr9_-_91793681
|
2.153
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr20_-_14318200
|
2.134
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr4_-_38806410
|
2.133
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr13_-_48987247
|
2.131
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr11_+_113930296
|
2.125
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr4_-_100273831
|
2.113
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr11_+_20049128
|
2.108
|
|
NAV2
|
neuron navigator 2
|
|
chr6_-_111136616
|
2.096
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr9_+_12774988
|
2.092
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr5_-_41510627
|
2.088
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr12_+_18891040
|
2.073
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr4_-_105416050
|
2.070
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr10_-_56560947
|
2.069
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr10_-_61122660
|
2.068
|
NM_001143773
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr6_-_11382501
|
2.066
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_-_221915426
|
2.064
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_+_51435911
|
2.063
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr17_-_4693595
|
2.056
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr2_-_145277879
|
2.047
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr4_-_1722978
|
2.040
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr14_-_23285011
|
2.031
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr7_+_139528951
|
2.026
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr12_-_42877785
|
2.024
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr3_+_89156673
|
2.019
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr2_-_60780767
|
2.017
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr6_-_39693110
|
2.007
|
NM_145027
|
KIF6
|
kinesin family member 6
|
|
chr5_+_125967405
|
2.007
|
NM_207408
|
C5orf48
|
chromosome 5 open reading frame 48
|
|
chr12_-_42877743
|
2.002
|
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr19_-_51845377
|
1.996
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr15_-_52970808
|
1.994
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr5_-_138210992
|
1.982
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr7_+_145813452
|
1.982
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr8_-_20112691
|
1.973
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr15_+_59903980
|
1.966
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr13_+_97928196
|
1.962
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr22_+_38201300
|
1.958
|
|
H1F0
|
H1 histone family, member 0
|
|
chr16_+_29823408
|
1.957
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr1_-_168698412
|
1.953
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr4_+_86851425
|
1.950
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr6_-_33168448
|
1.945
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr1_+_203734283
|
1.934
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr18_-_24445652
|
1.922
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr17_-_77478562
|
1.919
|
NM_001082575
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr17_-_48546174
|
1.914
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr14_-_22005035
|
1.908
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr12_-_86230158
|
1.907
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr9_+_116356407
|
1.903
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_-_61245363
|
1.870
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chr6_-_33168438
|
1.861
|
|
RXRB
|
retinoid X receptor, beta
|
|
chrX_-_106960268
|
1.856
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_154540300
|
1.836
|
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr5_+_140588267
|
1.827
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr5_+_53751430
|
1.823
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr13_+_97928414
|
1.809
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr18_+_61223392
|
1.807
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chr7_+_114055048
|
1.784
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr16_-_54319966
|
1.769
|
|
IRX3
|
iroquois homeobox 3
|
|
chr3_-_147122070
|
1.763
|
NM_001168378
|
ZIC4
|
Zic family member 4
|
|
chr22_+_38201232
|
1.761
|
|
H1F0
|
H1 histone family, member 0
|
|
chr10_-_104178569
|
1.757
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr11_-_805102
|
1.757
|
NM_145886
NM_145887
|
PIDD
|
p53-induced death domain protein
|
|
chr18_-_45663667
|
1.755
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr1_-_54199876
|
1.746
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr6_-_48036385
|
1.745
|
NM_001013732
NM_207499
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr6_-_152957985
|
1.739
|
NM_033071
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr2_+_183943511
|
1.732
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr19_-_56249739
|
1.720
|
NM_176820
|
NLRP9
|
NLR family, pyrin domain containing 9
|
|
chr20_-_44455932
|
1.719
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr9_-_14398981
|
1.716
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr7_+_114055171
|
1.705
|
|
FOXP2
|
forkhead box P2
|
|
chr12_+_14538232
|
1.701
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chrX_-_125300079
|
1.698
|
NM_001013628
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12-like 2
|
|
chr19_-_45663407
|
1.692
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr19_-_40324747
|
1.687
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr16_+_29823557
|
1.685
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr7_+_120629444
|
1.684
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr2_-_145275086
|
1.680
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr4_+_94750041
|
1.677
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chrX_-_83442906
|
1.669
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr12_-_14924017
|
1.668
|
NM_175054
|
HIST4H4
|
histone cluster 4, H4
|