|
chr2_-_224467049
|
3.875
|
|
SCG2
|
secretogranin II
|
|
chr17_-_39092714
|
3.524
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_-_224467142
|
3.018
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr2_-_224467095
|
3.005
|
|
SCG2
|
secretogranin II
|
|
chr7_+_37779995
|
2.823
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr10_+_123754141
|
2.584
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr6_-_24877453
|
2.357
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr19_-_17516383
|
1.997
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr7_-_122342965
|
1.933
|
NM_198085
|
RNF148
|
ring finger protein 148
|
|
chr2_-_31030271
|
1.910
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr4_-_184243578
|
1.778
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chrM_+_8857
|
1.701
|
|
|
|
|
chr20_-_44006876
|
1.671
|
NM_014477
|
TP53TG5
|
TP53 target 5
|
|
chr17_+_38465422
|
1.652
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr19_+_45174723
|
1.623
|
NM_001127893
NM_020219
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19
|
|
chr20_+_43803539
|
1.615
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr1_+_196621007
|
1.569
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr12_-_89746244
|
1.564
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr19_-_55668956
|
1.549
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr12_-_89745753
|
1.513
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr6_-_133078894
|
1.427
|
NM_001242350
NM_004665
|
VNN2
|
vanin 2
|
|
chr12_-_16757944
|
1.415
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_42908178
|
1.370
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr3_+_157154579
|
1.369
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chrM_-_9203
|
1.294
|
|
|
|
|
chr16_-_72128214
|
1.263
|
NM_001142318
|
TXNL4B
|
thioredoxin-like 4B
|
|
chr20_-_34243118
|
1.254
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr11_-_102714240
|
1.241
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chrM_+_9182
|
1.218
|
|
|
|
|
chr9_-_112970412
|
1.218
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr12_-_89746238
|
1.215
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr19_+_48949029
|
1.212
|
NM_031485
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr2_+_102608305
|
1.167
|
NM_004633
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr1_-_110155672
|
1.162
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chrM_+_3920
|
1.156
|
|
|
|
|
chr1_-_153067000
|
1.148
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr19_+_37180301
|
1.133
|
NM_152603
|
ZNF567
|
zinc finger protein 567
|
|
chrM_+_8364
|
1.133
|
|
|
|
|
chr2_+_228678556
|
1.126
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr7_+_112063131
|
1.118
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chrM_+_9217
|
1.107
|
|
|
|
|
chr11_-_128457452
|
1.103
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_153044083
|
1.061
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr5_+_159614373
|
1.057
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr2_+_234590583
|
1.046
|
NM_019077
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7
|
|
chr10_-_82049178
|
1.046
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr19_-_6670598
|
1.041
|
NM_003807
NM_172014
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr11_+_66742747
|
1.036
|
NM_001136485
|
C11orf86
|
chromosome 11 open reading frame 86
|
|
chr19_+_48325098
|
1.006
|
NM_000554
|
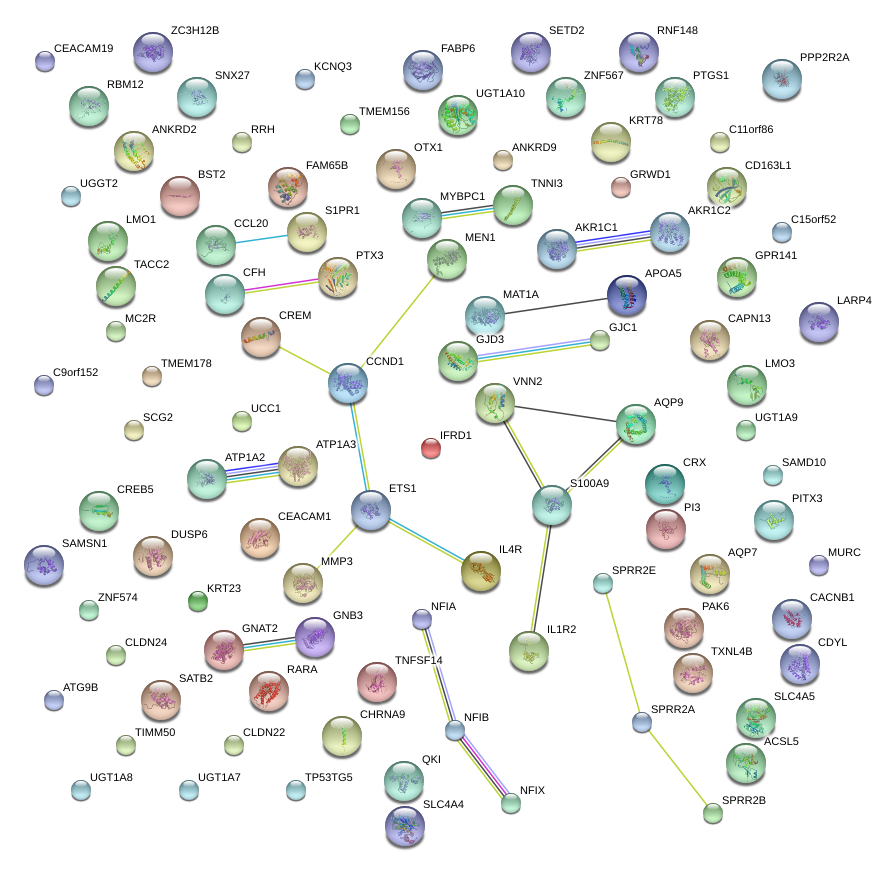
CRX
|
cone-rod homeobox
|
|
chr1_+_160085519
|
1.003
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr10_+_5005685
|
0.997
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr10_+_99332197
|
0.994
|
NM_001129981
NM_020349
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle)
|
|
chr12_-_7596722
|
0.975
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr8_+_26150731
|
0.964
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr8_-_133459508
|
0.963
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr12_+_101988746
|
0.963
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr12_+_6949374
|
0.960
|
NM_002075
|
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3
|
|
chrM_-_379
|
0.948
|
|
|
|
|
chr12_-_89745997
|
0.940
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr4_+_110749149
|
0.926
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chr15_+_58430394
|
0.924
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr18_-_13915534
|
0.915
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr2_-_74570533
|
0.912
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr12_-_53242716
|
0.902
|
NM_173352
|
KRT78
|
keratin 78
|
|
chr16_+_27341305
|
0.896
|
|
IL4R
|
interleukin 4 receptor
|
|
chr19_+_42580333
|
0.869
|
|
ZNF574
|
zinc finger protein 574
|
|
chr1_+_61547388
|
0.867
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_39033981
|
0.855
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr6_+_163984574
|
0.855
|
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr9_+_103340335
|
0.855
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr2_-_200325200
|
0.853
|
|
SATB2
|
SATB homeobox 2
|
|
chr11_-_116663106
|
0.844
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr11_+_69467210
|
0.839
|
|
CCND1
|
cyclin D1
|
|
chr19_-_43032525
|
0.827
|
NM_001024912
NM_001184813
NM_001184815
NM_001184816
NM_001205344
NM_001712
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr19_+_39971051
|
0.823
|
NM_001001563
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr12_+_50869531
|
0.818
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr10_+_114154522
|
0.818
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr2_+_63277191
|
0.798
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_+_153330329
|
0.792
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr4_+_40337389
|
0.790
|
NM_017581
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9
|
|
chr20_-_62611308
|
0.781
|
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr21_-_15918635
|
0.775
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr10_+_35484054
|
0.764
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr7_+_28530747
|
0.762
|
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_+_101702561
|
0.760
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr7_-_150721551
|
0.757
|
NM_173681
|
ATG9B
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chrX_+_64708614
|
0.748
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr15_-_40633122
|
0.736
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr2_+_39893021
|
0.735
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr15_+_40509628
|
0.733
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr1_+_151611331
|
0.733
|
|
SNX27
|
sorting nexin family member 27
|
|
chr9_+_125137600
|
0.733
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr22_+_22676814
|
0.724
|
|
|
|
|
chr6_+_4890225
|
0.713
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chr2_+_234580504
|
0.710
|
NM_021027
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr17_-_37350051
|
0.707
|
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr3_-_47098906
|
0.701
|
|
SETD2
|
SET domain containing 2
|
|
chr3_+_44692500
|
0.696
|
|
ZNF35
|
zinc finger protein 35
|
|
chr3_+_158787040
|
0.689
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr10_-_5045967
|
0.688
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr16_-_55866972
|
0.686
|
|
CES1
|
carboxylesterase 1
|
|
chr7_-_128415843
|
0.681
|
NM_001708
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive
|
|
chr9_+_38392658
|
0.680
|
NM_000692
|
ALDH1B1
|
aldehyde dehydrogenase 1 family, member B1
|
|
chr11_-_102668892
|
0.676
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr9_-_71590754
|
0.674
|
|
|
|
|
chrM_+_10704
|
0.653
|
|
|
|
|
chr4_+_95376396
|
0.651
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr10_+_124030820
|
0.651
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chr9_-_104198045
|
0.647
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr12_-_50616425
|
0.646
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr6_+_7107799
|
0.645
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr11_-_67141008
|
0.644
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr7_+_5013618
|
0.635
|
|
RNF216P1
|
ring finger protein 216 pseudogene 1
|
|
chr17_-_38859976
|
0.629
|
NM_019016
|
KRT24
|
keratin 24
|
|
chrX_+_53123211
|
0.628
|
|
LOC100132984
|
uncharacterized LOC100132984
|
|
chr9_+_38392696
|
0.628
|
|
ALDH1B1
|
aldehyde dehydrogenase 1 family, member B1
|
|
chr17_-_39280377
|
0.625
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr19_+_45349392
|
0.624
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr16_+_22524843
|
0.623
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr20_+_4702499
|
0.621
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr1_-_39347258
|
0.619
|
NM_030772
|
GJA9-MYCBP
GJA9
|
GJA9-MYCBP readthrough
gap junction protein, alpha 9, 59kDa
|
|
chr9_+_125132823
|
0.617
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr18_+_33234532
|
0.614
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr9_+_125132785
|
0.606
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr12_+_112890999
|
0.602
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr11_-_102962903
|
0.600
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chrM_+_11427
|
0.599
|
|
|
|
|
chr10_+_5005453
|
0.599
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr9_+_131445933
|
0.591
|
NM_001122821
|
SET
|
SET nuclear oncogene
|
|
chr8_+_39771327
|
0.591
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr16_-_75282137
|
0.591
|
NM_001170719
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr4_-_159644439
|
0.582
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr2_-_217560247
|
0.581
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr14_+_21510384
|
0.579
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr12_-_11175169
|
0.579
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr9_+_38392700
|
0.573
|
|
ALDH1B1
|
aldehyde dehydrogenase 1 family, member B1
|
|
chr7_+_129269916
|
0.570
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr10_-_101690655
|
0.565
|
|
DNMBP
|
dynamin binding protein
|
|
chr1_-_28520379
|
0.565
|
NM_001164721
NM_001164722
|
PTAFR
|
platelet-activating factor receptor
|
|
chr13_-_99910548
|
0.564
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chrX_+_41548225
|
0.562
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr19_+_42580275
|
0.561
|
NM_022752
|
ZNF574
|
zinc finger protein 574
|
|
chr16_+_4545858
|
0.559
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr11_+_101983170
|
0.558
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr6_-_30899926
|
0.552
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr10_-_124605690
|
0.552
|
NM_022034
|
CUZD1
|
CUB and zona pellucida-like domains 1
|
|
chr12_-_52761254
|
0.551
|
NM_002283
|
KRT85
|
keratin 85
|
|
chr21_+_45209448
|
0.550
|
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr5_+_70944968
|
0.543
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr19_-_5838740
|
0.537
|
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr5_-_148442606
|
0.536
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr17_-_48207113
|
0.536
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr2_+_234545099
|
0.535
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr11_+_36317724
|
0.533
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr6_-_138893666
|
0.531
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr11_+_61717355
|
0.530
|
NM_001139443
NM_004183
|
BEST1
|
bestrophin 1
|
|
chr1_-_109849613
|
0.530
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr11_+_112047088
|
0.530
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr12_-_49449106
|
0.530
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr2_-_89513056
|
0.530
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr10_+_28878666
|
0.529
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr2_-_96947640
|
0.529
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr22_+_32753853
|
0.529
|
NM_001098535
|
RFPL3
|
ret finger protein-like 3
|
|
chrX_-_110655384
|
0.527
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr10_+_5005601
|
0.526
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_+_156863522
|
0.526
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr16_-_90076528
|
0.526
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr1_+_24834083
|
0.523
|
NM_001251978
|
RCAN3
|
RCAN family member 3
|
|
chr2_-_183387063
|
0.522
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr12_+_110205816
|
0.521
|
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr19_+_54376975
|
0.520
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr5_+_43609261
|
0.520
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr2_+_211342405
|
0.509
|
NM_001122633
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr13_-_46756291
|
0.500
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr2_-_89619820
|
0.494
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr10_-_118680978
|
0.493
|
|
KIAA1598
|
KIAA1598
|
|
chr20_-_3762060
|
0.493
|
NM_015417
|
SPEF1
|
sperm flagellar 1
|
|
chr3_-_61728810
|
0.490
|
|
|
|
|
chrX_+_110339374
|
0.488
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr9_+_4839763
|
0.487
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr1_-_156786500
|
0.485
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr12_+_72332625
|
0.483
|
NM_173353
|
TPH2
|
tryptophan hydroxylase 2
|
|
chr12_+_56915587
|
0.483
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr1_+_161195832
|
0.483
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr2_+_228356287
|
0.481
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr8_-_38205652
|
0.480
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr5_+_36608430
|
0.476
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr14_-_23540770
|
0.475
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chrM_+_2211
|
0.474
|
|
|
|
|
chr19_-_633526
|
0.473
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr5_+_132432880
|
0.472
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chrX_-_80457440
|
0.472
|
NM_030763
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr15_-_75194542
|
0.472
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr22_-_36681757
|
0.469
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr1_+_27648746
|
0.468
|
|
LOC644961
TMEM222
|
actin, beta pseudogene
transmembrane protein 222
|
|
chrM_+_9725
|
0.468
|
|
|
|
|
chr12_+_53645369
|
0.463
|
NM_001170790
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr16_-_31076408
|
0.461
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr7_+_116660263
|
0.461
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr2_-_55237255
|
0.460
|
NM_007008
|
RTN4
|
reticulon 4
|