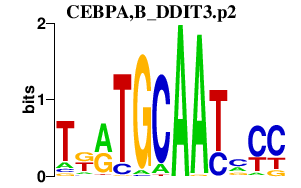
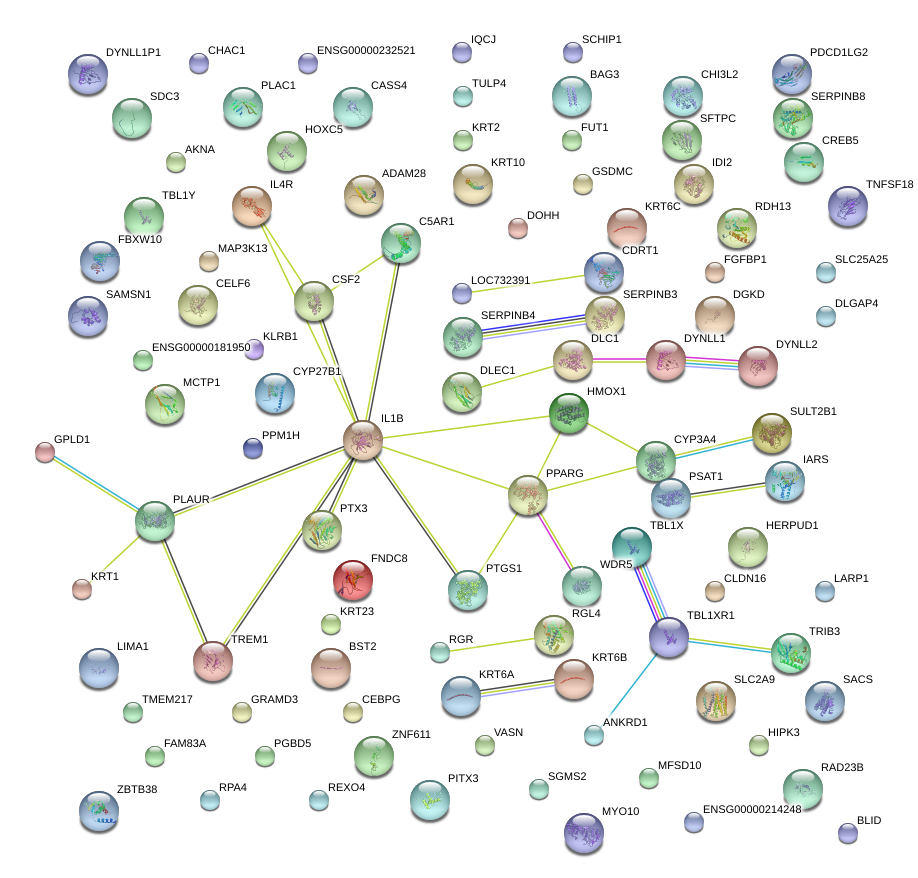
|
chr17_-_39092714
|
20.226
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr19_-_44174497
|
5.456
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr12_-_53074172
|
5.177
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr10_-_92680784
|
4.831
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr10_-_92681025
|
3.845
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_-_71590754
|
3.157
|
|
|
|
|
chr18_+_61637262
|
3.010
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chrX_+_9431314
|
2.930
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_-_230513390
|
2.888
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr19_-_49258646
|
2.861
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr12_-_52845862
|
2.779
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr3_+_190105660
|
2.597
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr9_+_130860769
|
2.337
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr7_+_28338939
|
2.335
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_-_113594325
|
2.285
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr4_+_108814419
|
2.194
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr12_-_63328857
|
1.979
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr1_+_111770280
|
1.656
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr17_-_38978824
|
1.647
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr2_+_234296799
|
1.641
|
NM_003648
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr4_-_10023094
|
1.555
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr12_-_52867521
|
1.533
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr15_+_41245635
|
1.487
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr3_+_157154579
|
1.450
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr6_+_158734902
|
1.421
|
|
|
|
|
chrX_-_133792497
|
1.401
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr1_-_23342342
|
1.363
|
NM_001242521
|
LOC729059
|
uncharacterized LOC729059
|
|
chr16_+_27341305
|
1.318
|
|
IL4R
|
interleukin 4 receptor
|
|
chr12_+_54426831
|
1.314
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr10_-_29923841
|
1.291
|
|
|
|
|
chr19_-_55580896
|
1.279
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr8_-_130799133
|
1.254
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr3_-_42843217
|
1.247
|
|
|
|
|
chr19_+_49078992
|
1.220
|
NM_004605
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr19_+_47813074
|
1.195
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr4_-_15940362
|
1.190
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr17_-_15523017
|
1.157
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr7_+_28725597
|
1.123
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_167956545
|
1.123
|
|
|
|
|
chr12_+_65224214
|
1.096
|
|
|
|
|
chr18_-_61311465
|
1.074
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr5_+_125758818
|
1.040
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr6_-_41254392
|
1.011
|
NM_001242589
NM_001242590
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr6_+_158734476
|
1.007
|
|
TULP4
|
tubby like protein 4
|
|
chr12_-_50595303
|
0.981
|
NM_001243775
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr2_+_12858363
|
0.935
|
|
|
|
|
chr9_-_117156684
|
0.903
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr12_-_53045947
|
0.898
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr5_-_94224601
|
0.862
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr19_-_17516383
|
0.857
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr9_+_5510544
|
0.854
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr19_+_33864574
|
0.838
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr20_+_361938
|
0.835
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr17_+_18647325
|
0.834
|
NM_031456
|
FBXW10
|
F-box and WD repeat domain containing 10
|
|
chr5_+_131409484
|
0.812
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr1_-_31345361
|
0.803
|
|
SDC3
|
syndecan 3
|
|
chr10_-_1071797
|
0.800
|
NM_033261
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2
|
|
chr9_-_95055913
|
0.797
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr8_+_124194751
|
0.790
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr8_+_24151579
|
0.784
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr9_-_95055966
|
0.783
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr9_-_95055986
|
0.782
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr20_+_54987167
|
0.769
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr11_+_33363104
|
0.760
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr9_+_110045551
|
0.757
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr8_+_22019167
|
0.728
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr8_+_124194873
|
0.718
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr9_+_130860816
|
0.708
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr3_+_141043054
|
0.706
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr9_-_95055989
|
0.693
|
NM_002161
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr4_-_2935650
|
0.684
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr9_+_110045516
|
0.678
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr8_-_12973752
|
0.677
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_-_37225360
|
0.676
|
NM_145316
|
TMEM217
|
transmembrane protein 217
|
|
chr13_-_24007822
|
0.675
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr5_+_154135029
|
0.665
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr21_-_15918635
|
0.665
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr9_+_80911990
|
0.662
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr1_-_173020011
|
0.658
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr12_-_58160823
|
0.658
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr3_+_12392993
|
0.654
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_+_86004808
|
0.645
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr17_+_33448597
|
0.634
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr16_-_30787298
|
0.631
|
|
|
|
|
chr19_-_53233134
|
0.627
|
NM_030972
|
ZNF611
|
zinc finger protein 611
|
|
chr22_+_35779103
|
0.627
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr15_-_72598655
|
0.608
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr9_+_137004913
|
0.600
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr3_+_158787040
|
0.579
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr7_-_104567089
|
0.570
|
|
LOC723809
|
uncharacterized LOC723809
|
|
chr19_-_3500937
|
0.569
|
NM_001145165
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase
|
|
chr9_+_110045565
|
0.560
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr11_-_121986800
|
0.559
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr16_+_56966025
|
0.559
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr6_-_24489738
|
0.559
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr10_+_121410751
|
0.549
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr20_+_34995443
|
0.546
|
NM_014902
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chrX_+_96138906
|
0.542
|
NM_013347
|
RPA4
|
replication protein A4, 30kDa
|
|
chr9_+_125132823
|
0.541
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr3_+_185080835
|
0.537
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr5_-_16738346
|
0.536
|
|
MYO10
|
myosin X
|
|
chr16_+_4430878
|
0.535
|
|
VASN
|
vasorin
|
|
chr6_+_167704802
|
0.534
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr12_-_121736110
|
0.520
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr19_+_33865429
|
0.513
|
NM_001252296
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr20_-_47874147
|
0.512
|
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr10_-_116444374
|
0.506
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_-_116444112
|
0.500
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_+_11674241
|
0.496
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr5_-_176836488
|
0.494
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr4_-_2935851
|
0.492
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr21_-_11098819
|
0.489
|
NM_001187
NM_182484
NM_181704
NM_182481
NM_182482
|
BAGE
BAGE5
BAGE4
BAGE3
BAGE2
|
B melanoma antigen
B melanoma antigen family, member 5
B melanoma antigen family, member 4
B melanoma antigen family, member 3
B melanoma antigen family, member 2
|
|
chr5_-_66492574
|
0.489
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr10_+_69869249
|
0.487
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr5_+_161275897
|
0.486
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_-_131630939
|
0.482
|
|
|
|
|
chr3_+_48264836
|
0.479
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr6_-_33548017
|
0.475
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr18_-_55289026
|
0.470
|
NM_004539
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr21_+_30400229
|
0.464
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr17_-_77813049
|
0.461
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr4_+_95376396
|
0.459
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr15_+_89182038
|
0.453
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr12_+_75874512
|
0.445
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr18_-_55289007
|
0.437
|
|
NARS
|
asparaginyl-tRNA synthetase
|
|
chr1_-_153283166
|
0.435
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr16_+_67381252
|
0.435
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr20_+_361675
|
0.433
|
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr19_-_47128293
|
0.430
|
NM_000960
|
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP)
|
|
chr1_-_153521717
|
0.425
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr6_-_42162693
|
0.423
|
NM_002098
|
GUCA1B
|
guanylate cyclase activator 1B (retina)
|
|
chr11_-_104972147
|
0.420
|
NM_001007232
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CARD17
CASP1
|
caspase recruitment domain family, member 17
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr19_+_48949029
|
0.415
|
NM_031485
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr11_+_63304272
|
0.415
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr1_+_153330329
|
0.413
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr16_+_3405888
|
0.413
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr17_-_30228750
|
0.413
|
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast)
|
|
chr3_+_186435097
|
0.411
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr3_-_187420344
|
0.407
|
NM_001004312
|
RTP2
|
receptor (chemosensory) transporter protein 2
|
|
chr19_+_13250072
|
0.398
|
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr9_+_684193
|
0.396
|
|
|
|
|
chrM_-_9203
|
0.393
|
|
|
|
|
chr9_-_130712866
|
0.393
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr11_-_66675334
|
0.379
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr3_+_156394452
|
0.371
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr5_-_16742336
|
0.371
|
|
MYO10
|
myosin X
|
|
chr7_-_91763677
|
0.370
|
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr2_+_237993954
|
0.367
|
NM_006710
NM_198189
|
COPS8
|
COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis)
|
|
chr17_-_18177716
|
0.366
|
|
TOP3A
|
topoisomerase (DNA) III alpha
|
|
chr17_+_20771745
|
0.366
|
|
|
|
|
chr12_-_113841654
|
0.362
|
NM_006843
|
SDS
|
serine dehydratase
|
|
chr1_-_161207879
|
0.358
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr16_-_21663962
|
0.355
|
NM_005849
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr6_+_30297087
|
0.354
|
NM_001199119
|
TRIM39-RPP21
TRIM39
|
TRIM39-RPP21 readthrough
tripartite motif containing 39
|
|
chr14_+_35514112
|
0.349
|
NM_001079519
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr9_+_125132785
|
0.348
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr16_+_56815703
|
0.347
|
NM_001242795
|
NUP93
|
nucleoporin 93kDa
|
|
chr17_-_7166505
|
0.346
|
NM_001185022
|
CLDN7
|
claudin 7
|
|
chr12_-_57009361
|
0.345
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr12_+_113344811
|
0.343
|
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr10_+_70980058
|
0.341
|
NM_025130
|
HKDC1
|
hexokinase domain containing 1
|
|
chr19_+_37180301
|
0.340
|
NM_152603
|
ZNF567
|
zinc finger protein 567
|
|
chr3_-_47957475
|
0.340
|
|
|
|
|
chr15_-_51386815
|
0.332
|
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr10_-_50396380
|
0.329
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chrX_-_40506800
|
0.328
|
NM_144970
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr11_-_65547821
|
0.325
|
NM_138368
|
DKFZp761E198
|
adaptor protein 5
|
|
chr4_+_184020343
|
0.322
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr3_-_49399969
|
0.321
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr14_+_29241989
|
0.320
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr4_+_41700736
|
0.316
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr21_-_47604360
|
0.315
|
NM_001142854
NM_032261
|
C21orf56
|
chromosome 21 open reading frame 56
|
|
chr9_+_116342939
|
0.308
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr5_-_176836535
|
0.303
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr12_-_48744553
|
0.302
|
NM_001172681
NM_152320
|
ZNF641
|
zinc finger protein 641
|
|
chr20_+_24929904
|
0.301
|
|
CST7
|
cystatin F (leukocystatin)
|
|
chr5_+_125759076
|
0.300
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr6_+_12717036
|
0.296
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr2_-_38525527
|
0.293
|
|
ATL2
|
atlastin GTPase 2
|
|
chr3_+_159706622
|
0.292
|
NM_000882
|
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35)
|
|
chr4_+_113739203
|
0.291
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr22_+_45072687
|
0.291
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr9_-_95186559
|
0.290
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr4_-_71532341
|
0.287
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr15_-_34212523
|
0.286
|
|
|
|
|
chr2_-_230933618
|
0.285
|
NM_152527
|
SLC16A14
|
solute carrier family 16, member 14 (monocarboxylic acid transporter 14)
|
|
chr1_-_154842725
|
0.284
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr3_+_186330859
|
0.280
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr16_+_75252882
|
0.275
|
NM_001906
|
CTRB1
|
chymotrypsinogen B1
|
|
chr11_+_18287807
|
0.274
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chr7_-_91764036
|
0.273
|
NM_001146152
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr18_+_29598444
|
0.273
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr4_+_146031528
|
0.272
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr10_+_135160843
|
0.272
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr11_-_102668822
|
0.270
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr1_+_40859016
|
0.269
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr5_+_132009372
|
0.269
|
NM_000589
NM_172348
|
IL4
|
interleukin 4
|
|
chr3_+_186330711
|
0.268
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr6_-_11111955
|
0.268
|
NM_207582
|
ERVFRD-1
|
endogenous retrovirus group FRD, member 1
|
|
chr13_+_29598746
|
0.265
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|