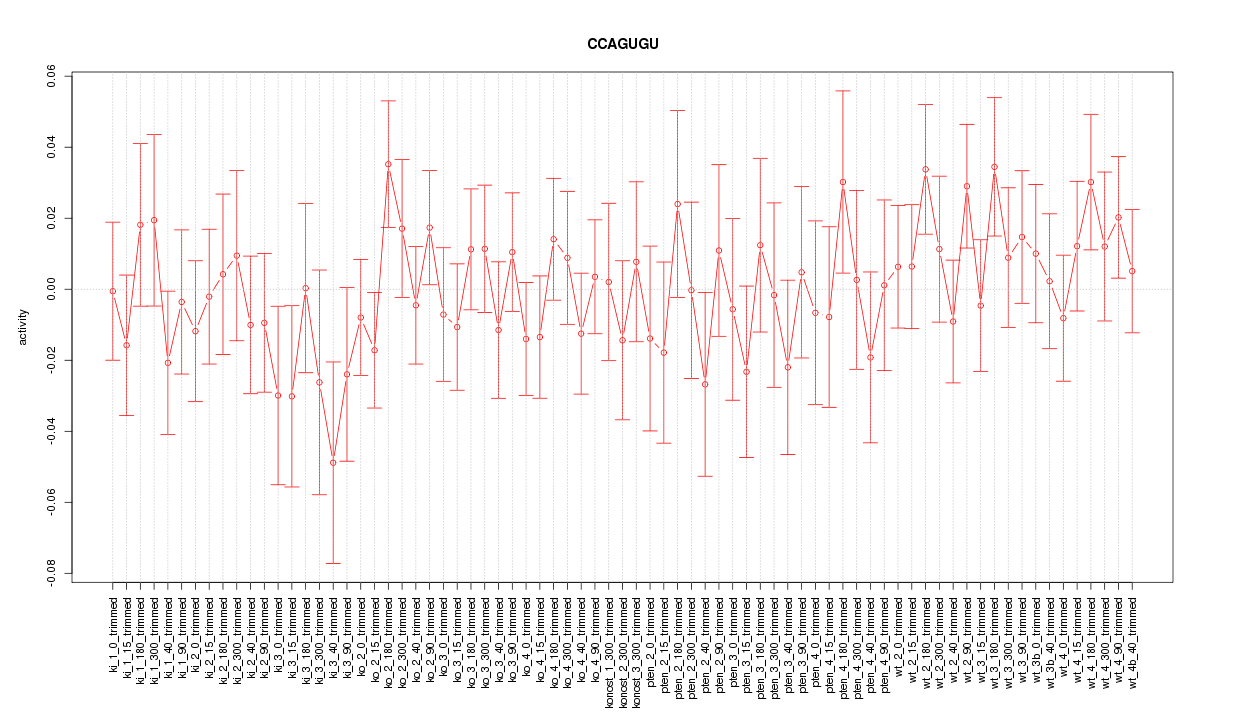
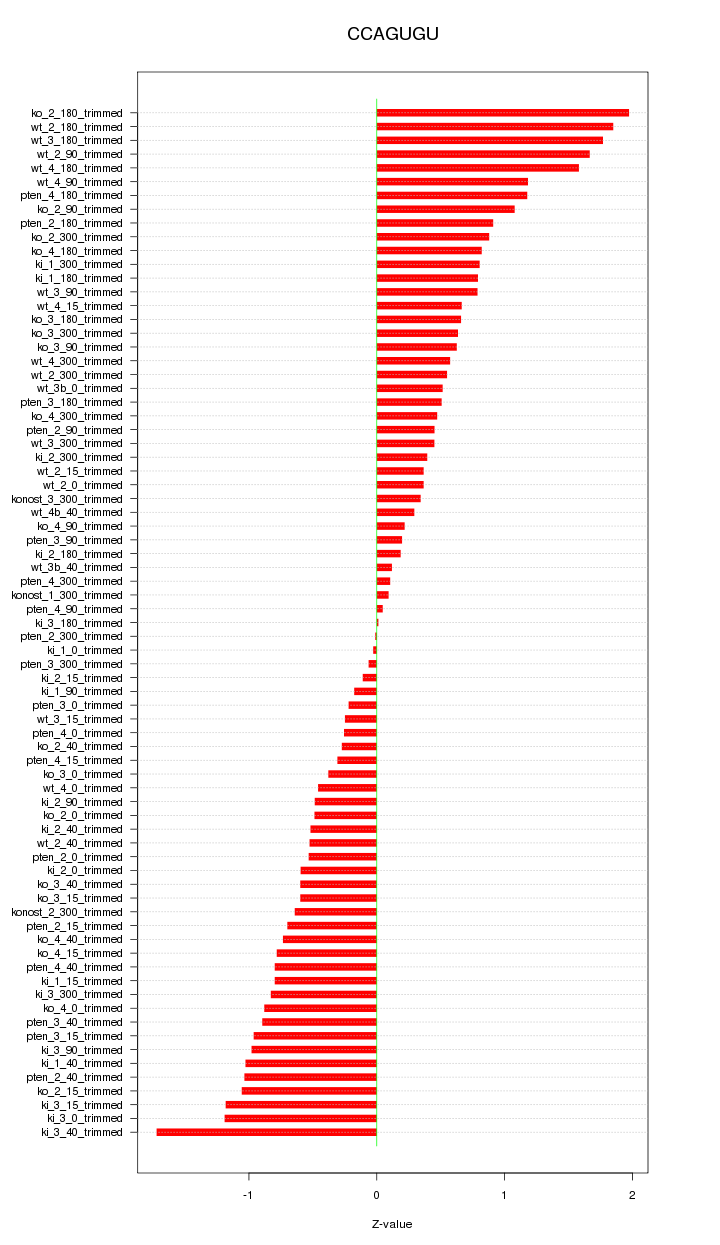
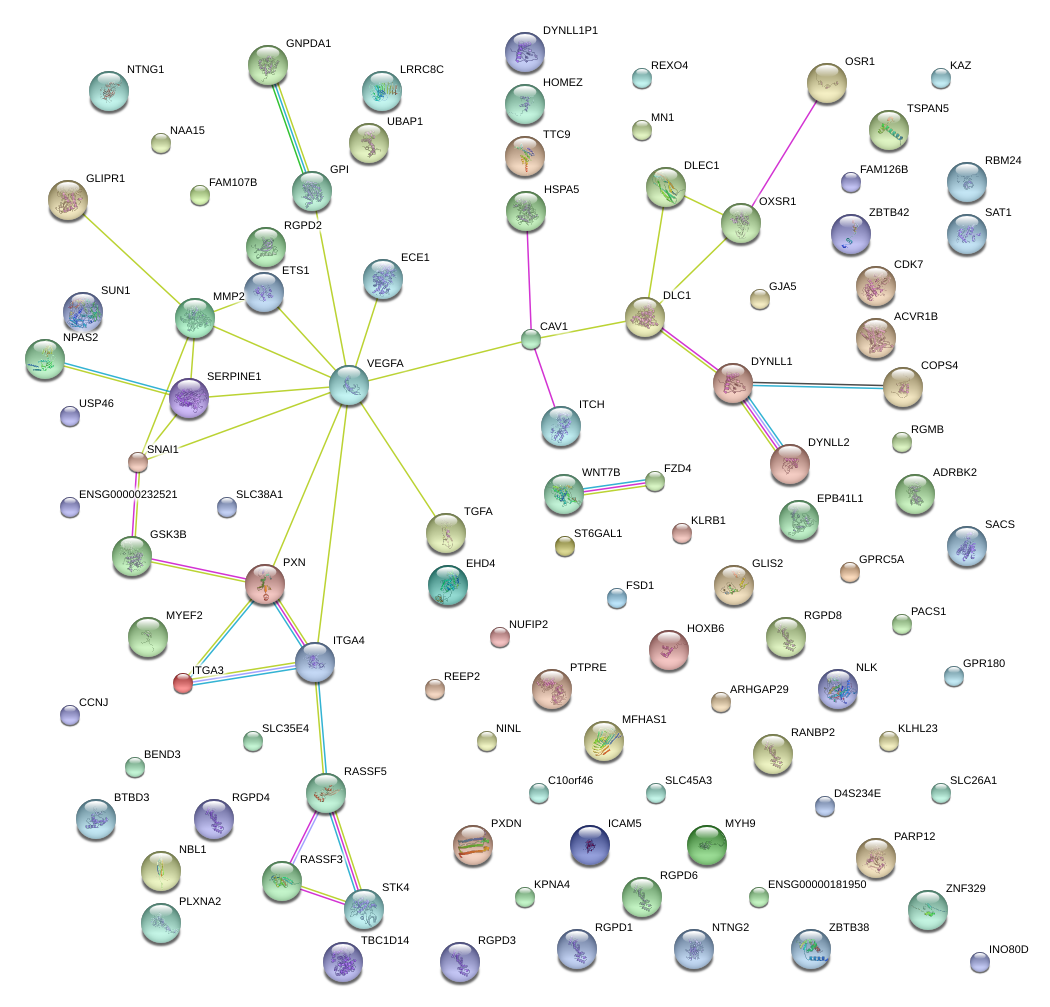
|
chr11_-_128457452
|
5.334
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr11_-_128392061
|
4.917
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr4_-_99579540
|
4.903
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr1_+_107682628
|
4.351
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr2_+_101436491
|
3.417
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr1_-_94703252
|
3.003
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr6_-_107435635
|
2.962
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr20_+_48599511
|
2.955
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr10_-_14816895
|
2.921
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr17_+_26369617
|
2.859
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr13_-_24007822
|
2.545
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr10_+_97803064
|
2.451
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr9_-_128003634
|
2.354
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr3_+_186739643
|
2.299
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr15_-_48470453
|
2.276
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr2_-_19558371
|
2.268
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr14_+_71108373
|
2.198
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr12_+_13043955
|
2.139
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr10_+_129845806
|
2.126
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr1_-_208417635
|
1.998
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr13_+_95254021
|
1.967
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr4_+_4387982
|
1.962
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr17_-_46682333
|
1.934
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr16_+_55512801
|
1.934
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr22_+_31031672
|
1.907
|
NM_001001479
|
SLC35E4
|
solute carrier family 35, member E4
|
|
chr12_+_52347163
|
1.876
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr3_+_186648273
|
1.853
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr10_+_129705299
|
1.781
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr17_+_48133331
|
1.750
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr3_-_160283361
|
1.733
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr6_+_17282931
|
1.718
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr8_-_8750739
|
1.678
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr19_+_4304590
|
1.665
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr15_-_42264749
|
1.656
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr8_-_12973752
|
1.622
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_-_147245483
|
1.591
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr22_-_46373007
|
1.575
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr1_-_21671905
|
1.540
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr12_+_65004191
|
1.537
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr20_-_25566137
|
1.532
|
NM_025176
|
NINL
|
ninein-like
|
|
chr2_+_170590241
|
1.522
|
NM_144711
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr7_+_100770378
|
1.514
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr6_+_17281773
|
1.469
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr5_+_98104998
|
1.431
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr4_+_140222668
|
1.422
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr12_-_120687956
|
1.410
|
NM_025157
|
PXN
|
paxillin
|
|
chr2_-_206950895
|
1.350
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr7_+_872137
|
1.334
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr1_+_90098643
|
1.312
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_-_147232684
|
1.277
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr19_-_58662144
|
1.266
|
NM_024620
|
ZNF329
|
zinc finger protein 329
|
|
chr12_+_75874512
|
1.252
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr5_+_137774632
|
1.214
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr5_+_68530621
|
1.206
|
NM_001799
|
CDK7
|
cyclin-dependent kinase 7
|
|
chr22_-_36784055
|
1.204
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr3_-_119812973
|
1.181
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr10_-_120514669
|
1.167
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr2_-_201936389
|
1.167
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr22_-_28197469
|
1.165
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr2_-_70781146
|
1.160
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr1_-_21605984
|
1.154
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr20_+_32951054
|
1.110
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr20_+_43595119
|
1.087
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr20_+_11898536
|
1.079
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr4_-_53522756
|
1.075
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr14_+_105266878
|
1.068
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr9_+_34179002
|
1.056
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr4_+_6988888
|
1.053
|
NM_001113363
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr2_+_109335906
|
1.050
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr22_+_25960738
|
1.044
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr2_+_182321618
|
1.029
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr7_-_139763520
|
1.023
|
NM_022750
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr7_+_116164838
|
1.016
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr16_+_55515468
|
1.015
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr6_+_43737937
|
1.007
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_-_205649616
|
1.007
|
NM_033102
|
SLC45A3
|
solute carrier family 45, member 3
|
|
chr11_-_86666211
|
0.987
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr16_+_4382224
|
0.986
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr7_+_116165043
|
0.983
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr1_+_19969722
|
0.977
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr3_+_141043054
|
0.970
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr20_+_34700257
|
0.962
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr1_-_21616648
|
0.951
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr17_-_27621163
|
0.941
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chrX_+_23801274
|
0.939
|
NM_002970
|
SAT1
|
spermidine/spermine N1-acetyltransferase 1
|
|
chr11_-_65381641
|
0.936
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr5_+_156887026
|
0.933
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr12_-_80307690
|
0.927
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr2_+_99953749
|
0.920
|
NM_015904
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr12_-_51477299
|
0.920
|
NM_030809
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr17_+_74261280
|
0.919
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr5_-_179229874
|
0.909
|
NM_054013
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr9_+_6215785
|
0.902
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr12_+_50794591
|
0.887
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr6_+_20402040
|
0.886
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr19_+_11094799
|
0.871
|
NM_001128845
NM_001128846
NM_001128847
NM_001128848
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr8_-_12990731
|
0.866
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_19972259
|
0.853
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr8_-_13372183
|
0.853
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr11_-_102323353
|
0.846
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr15_+_80987609
|
0.843
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr1_+_244216506
|
0.834
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr17_-_15168590
|
0.833
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr7_+_116166346
|
0.825
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr2_-_47168902
|
0.822
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr9_+_132962871
|
0.814
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr5_-_58882274
|
0.806
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_70820363
|
0.796
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chrX_+_49644469
|
0.794
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr2_-_47143250
|
0.793
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr16_+_31484497
|
0.792
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_-_21616888
|
0.783
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr10_-_62332666
|
0.773
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_118501981
|
0.767
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr1_-_19282773
|
0.758
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr12_-_80329234
|
0.753
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr14_+_67999915
|
0.746
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chrX_+_48916515
|
0.744
|
NM_001163322
NM_033626
NM_001163321
NM_001163323
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr12_-_120703549
|
0.740
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr1_-_145826928
|
0.733
|
NM_001097612
NM_001097613
|
GPR89B
GPR89A
|
G protein-coupled receptor 89B
G protein-coupled receptor 89A
|
|
chr1_+_33004771
|
0.731
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr9_+_131445933
|
0.729
|
NM_001122821
|
SET
|
SET nuclear oncogene
|
|
chr3_+_63953419
|
0.724
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr3_-_52713728
|
0.722
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr1_-_115212628
|
0.716
|
NM_198459
|
DENND2C
|
DENN/MADD domain containing 2C
|
|
chrX_+_117480027
|
0.709
|
NM_001184965
NM_001184966
NM_019045
|
WDR44
|
WD repeat domain 44
|
|
chr1_+_19970207
|
0.701
|
NM_001204084
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr6_-_38563763
|
0.695
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr12_+_57482674
|
0.690
|
NM_005967
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2)
|
|
chr2_+_219472487
|
0.685
|
NM_032726
|
PLCD4
|
phospholipase C, delta 4
|
|
chr1_+_19971865
|
0.679
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr4_+_6911094
|
0.677
|
NM_001113361
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr10_-_105614952
|
0.675
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_+_36583369
|
0.674
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr1_+_244214552
|
0.671
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_47142950
|
0.668
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr19_+_7599590
|
0.663
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr11_+_119076974
|
0.659
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr3_+_63884074
|
0.659
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr5_-_59783885
|
0.646
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr14_+_62162042
|
0.644
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr7_-_21985421
|
0.639
|
NM_001127370
NM_001127371
NM_018719
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr1_-_182573393
|
0.639
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr20_-_10654429
|
0.638
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr11_-_119599253
|
0.637
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr17_+_27717942
|
0.634
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr3_+_88188261
|
0.632
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_+_218458628
|
0.630
|
NM_016052
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae)
|
|
chr8_-_81787015
|
0.619
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr10_+_99498233
|
0.619
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr8_+_144680073
|
0.617
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr11_-_27528298
|
0.616
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chr19_+_7599037
|
0.616
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr16_+_31483066
|
0.616
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr4_+_25314392
|
0.608
|
NM_024936
|
ZCCHC4
|
zinc finger, CCHC domain containing 4
|
|
chr2_-_153573858
|
0.606
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr1_+_147400498
|
0.600
|
NM_016334
|
GPR89B
GPR89A
|
G protein-coupled receptor 89B
G protein-coupled receptor 89A
|
|
chr10_-_32217762
|
0.600
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr19_-_52531517
|
0.599
|
NM_025040
|
ZNF614
|
zinc finger protein 614
|
|
chr12_+_11802726
|
0.599
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr13_-_50510410
|
0.598
|
NM_001127482
NM_020456
|
SPRYD7
|
SPRY domain containing 7
|
|
chr12_-_80328926
|
0.598
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chrX_-_99891690
|
0.595
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr3_-_113415313
|
0.592
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr3_-_189840225
|
0.591
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr4_-_53525316
|
0.591
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr17_+_61086897
|
0.590
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr19_+_45349392
|
0.589
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr8_+_104310660
|
0.584
|
NM_001164615
|
FZD6
|
frizzled family receptor 6
|
|
chr3_-_172428773
|
0.578
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr5_-_58571916
|
0.578
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_56084656
|
0.576
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr19_-_16683188
|
0.573
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr17_+_7452374
|
0.564
|
NM_172089
NM_003809
|
TNFSF12-TNFSF13
TNFSF12
|
TNFSF12-TNFSF13 readthrough
tumor necrosis factor (ligand) superfamily, member 12
|
|
chr3_-_189838907
|
0.559
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr8_+_104311058
|
0.559
|
NM_001164616
NM_003506
|
FZD6
|
frizzled family receptor 6
|
|
chr15_-_56035176
|
0.549
|
NM_173814
|
PRTG
|
protogenin
|
|
chr15_-_83953465
|
0.542
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr17_-_35656691
|
0.541
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr8_-_91997437
|
0.540
|
NM_001190972
|
LOC100127983
|
uncharacterized LOC100127983
|
|
chr19_-_10121041
|
0.532
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr11_-_102962903
|
0.529
|
NM_032299
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr6_+_21593963
|
0.525
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr4_+_6911456
|
0.525
|
NM_020773
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr7_-_105162551
|
0.520
|
NM_019042
|
PUS7
|
pseudouridylate synthase 7 homolog (S. cerevisiae)
|
|
chr15_+_90544608
|
0.516
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr13_-_74707913
|
0.513
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_+_28995214
|
0.510
|
NM_006582
NM_024482
|
GMEB1
|
glucocorticoid modulatory element binding protein 1
|
|
chr2_+_210444364
|
0.510
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr17_-_79479831
|
0.509
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr10_-_121296044
|
0.506
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr6_+_52935770
|
0.499
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr5_+_139493689
|
0.498
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr3_-_125094003
|
0.497
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr16_+_66914380
|
0.497
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr2_+_191273012
|
0.496
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr19_-_56092184
|
0.495
|
NM_152600
|
ZNF579
|
zinc finger protein 579
|
|
chr3_+_112051914
|
0.495
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr12_+_52345437
|
0.494
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr10_-_71930185
|
0.484
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|