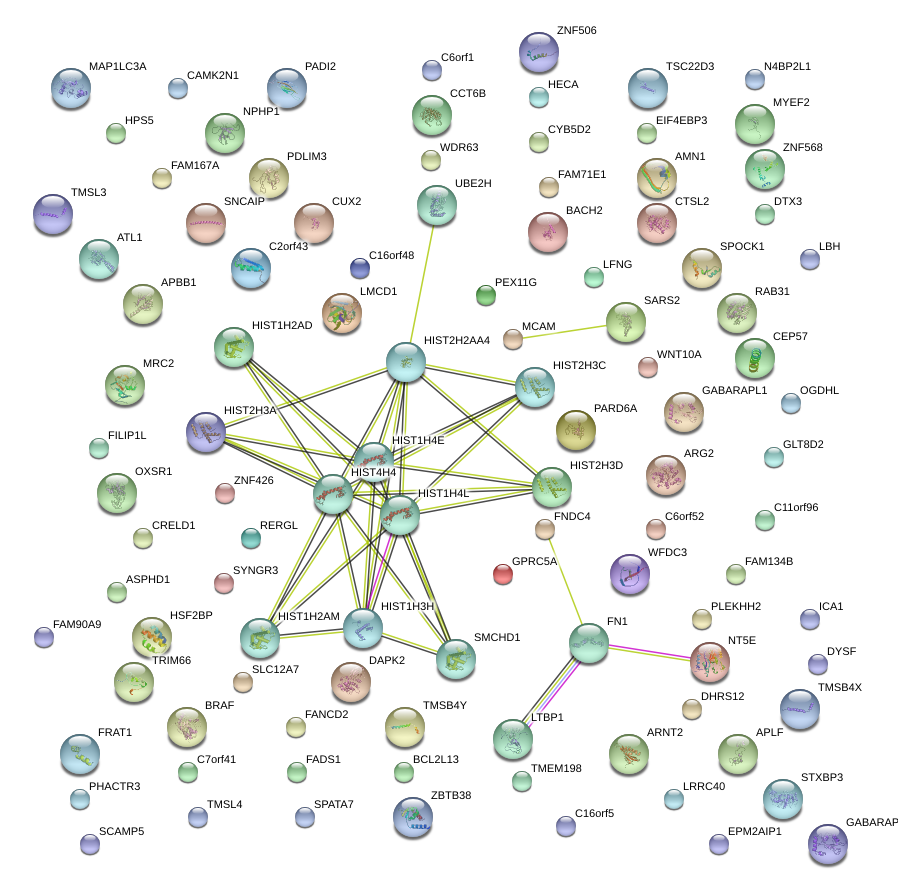
|
chr20_+_33146500
|
57.822
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr2_-_216300770
|
26.284
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr11_-_119187811
|
20.614
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr2_-_216300444
|
11.589
|
|
FN1
|
fibronectin 1
|
|
chr2_+_30454396
|
11.339
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr10_-_50970321
|
8.646
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr19_-_9649257
|
7.966
|
NM_024106
|
ZNF426
|
zinc finger protein 426
|
|
chr3_+_8543508
|
7.452
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr14_+_68086633
|
7.415
|
|
ARG2
|
arginase, type II
|
|
chr3_+_8543539
|
7.102
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr5_-_1112106
|
6.571
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr3_-_99594915
|
6.521
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_+_57998600
|
6.356
|
NM_178502
|
DTX3
|
deltex homolog 3 (Drosophila)
|
|
chr19_-_19932512
|
6.160
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr16_+_2039945
|
5.977
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr16_+_67694795
|
5.764
|
NM_001037281
NM_016948
|
PARD6A
|
par-6 partitioning defective 6 homolog alpha (C. elegans)
|
|
chr19_-_37407079
|
5.730
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr2_+_219745243
|
5.546
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr20_+_58179601
|
5.477
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr6_+_26204779
|
5.171
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr17_+_4046941
|
4.967
|
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr6_+_139456240
|
4.888
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr11_+_43963809
|
4.804
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr7_+_30174343
|
4.632
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr19_+_37407230
|
4.499
|
NM_001204835
NM_001204836
NM_001204837
NM_001204838
NM_001204839
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chrX_-_106960268
|
4.417
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_-_37406918
|
4.384
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr19_-_7553867
|
4.260
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chrX_-_106960028
|
4.168
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr4_-_186456580
|
4.168
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr5_+_121647773
|
4.154
|
NM_001242935
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr6_-_10694837
|
4.131
|
NM_001145020
|
C6orf52
|
chromosome 6 open reading frame 52
|
|
chr7_-_129592794
|
4.106
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr7_-_129592711
|
3.898
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr2_+_43864433
|
3.801
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr14_+_50999799
|
3.713
|
NM_001127713
|
ATL1
|
atlastin GTPase 1
|
|
chr15_-_48470319
|
3.593
|
|
MYEF2
|
myelin expression factor 2
|
|
chr2_+_220408742
|
3.576
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr14_+_68086655
|
3.465
|
|
ARG2
|
arginase, type II
|
|
chr9_-_99801365
|
3.465
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr11_-_6440631
|
3.350
|
NM_145689
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr16_-_67700571
|
3.237
|
NM_032140
|
C16orf48
|
chromosome 16 open reading frame 48
|
|
chr1_+_85527992
|
3.196
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr2_+_71680752
|
3.187
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr3_+_10068123
|
3.174
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr1_-_20812727
|
3.052
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr15_-_64338519
|
3.035
|
NM_014326
|
DAPK2
|
death-associated protein kinase 2
|
|
chr5_-_16509106
|
2.974
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr17_+_60704747
|
2.940
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr18_+_9708299
|
2.906
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_+_139927182
|
2.855
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr2_-_110962577
|
2.852
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr21_-_45079327
|
2.780
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr2_+_68694669
|
2.769
|
NM_173545
|
APLF
|
aprataxin and PNKP like factor
|
|
chr14_+_68086570
|
2.749
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr12_+_13044352
|
2.742
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr12_-_31881949
|
2.734
|
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr14_-_50329567
|
2.703
|
|
RN7SL2
|
RNA, 7SL, cytoplasmic 2
|
|
chr5_-_136834418
|
2.700
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr20_-_44420538
|
2.695
|
NM_080614
|
WFDC3
|
WAP four-disulfide core domain 3
|
|
chr5_-_16617085
|
2.681
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_-_50979461
|
2.662
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr12_+_10365415
|
2.636
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr15_+_75287829
|
2.629
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr10_+_99079017
|
2.613
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr6_+_86159738
|
2.586
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr11_-_61584451
|
2.575
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr2_-_27718098
|
2.575
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr2_-_27718059
|
2.524
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr14_+_88851987
|
2.499
|
NM_001040428
NM_018418
|
SPATA7
|
spermatogenesis associated 7
|
|
chr13_-_52378230
|
2.492
|
NM_001031719
NM_024705
|
DHRS12
|
dehydrogenase/reductase (SDR family) member 12
|
|
chr6_-_91006460
|
2.463
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_+_33172368
|
2.426
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr16_-_4588738
|
2.388
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_-_149812733
|
2.384
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr3_+_10068139
|
2.378
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr22_+_18121540
|
2.333
|
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr12_+_13044493
|
2.326
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr1_+_149824180
|
2.322
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr8_-_11324184
|
2.294
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr1_+_109289294
|
2.289
|
|
STXBP3
|
syntaxin binding protein 3
|
|
chr12_-_31882006
|
2.280
|
NM_001113402
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr7_-_140624280
|
2.259
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr1_+_201476283
|
2.255
|
|
|
|
|
chr14_-_103995336
|
2.218
|
|
|
|
|
chr1_-_17445865
|
2.198
|
NM_007365
|
PADI2
|
peptidyl arginine deiminase, type II
|
|
chr2_-_21022812
|
2.188
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr15_+_80696569
|
2.183
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr17_-_33288415
|
2.165
|
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2)
|
|
chr7_-_8301767
|
2.155
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr11_+_95523608
|
2.147
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr14_+_88852065
|
2.125
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr1_-_70671263
|
2.121
|
NM_017768
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr20_+_42839599
|
2.092
|
|
LOC100505783
|
uncharacterized LOC100505783
|
|
chr7_-_8301681
|
2.084
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr3_+_38206968
|
2.079
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr11_-_8680382
|
2.072
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr3_+_141087298
|
2.066
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr13_-_33002218
|
2.051
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr7_+_2557496
|
2.045
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr16_+_29912146
|
2.030
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr12_+_111471827
|
2.028
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chrX_+_12993224
|
2.014
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr5_-_136834231
|
2.005
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_-_18343670
|
1.988
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr3_+_9975513
|
1.953
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr6_+_27100784
|
1.951
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr8_+_26240522
|
1.939
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr1_+_52082755
|
1.939
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr17_+_57642885
|
1.918
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_+_109289266
|
1.900
|
NM_007269
|
STXBP3
|
syntaxin binding protein 3
|
|
chr6_+_86159301
|
1.896
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr14_+_23067128
|
1.885
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr8_-_66754276
|
1.884
|
|
PDE7A
|
phosphodiesterase 7A
|
|
chr14_-_50053068
|
1.877
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr5_-_72861208
|
1.866
|
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr16_-_4588379
|
1.863
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr1_+_109289345
|
1.855
|
|
STXBP3
|
syntaxin binding protein 3
|
|
chr5_+_60628085
|
1.844
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr6_-_91006580
|
1.832
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr22_+_18121446
|
1.824
|
NM_015367
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr6_-_53213699
|
1.813
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr3_+_183165402
|
1.812
|
|
LOC100505687
|
uncharacterized LOC100505687
|
|
chrX_+_43514157
|
1.811
|
|
MAOA
|
monoamine oxidase A
|
|
chr14_+_32030579
|
1.799
|
NM_025152
|
NUBPL
|
nucleotide binding protein-like
|
|
chr19_+_1104614
|
1.793
|
NM_001039848
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr17_+_33914320
|
1.782
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr8_-_71581419
|
1.772
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr13_+_32889594
|
1.769
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr6_-_116575170
|
1.763
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr11_-_6440288
|
1.756
|
NM_001164
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr2_+_43864405
|
1.756
|
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr12_+_56552141
|
1.751
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr20_+_19915716
|
1.736
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr4_+_170541728
|
1.710
|
|
CLCN3
|
chloride channel 3
|
|
chr5_-_121413906
|
1.706
|
|
LOX
|
lysyl oxidase
|
|
chr17_+_60705042
|
1.704
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr16_+_19567021
|
1.694
|
|
C16orf62
|
chromosome 16 open reading frame 62
|
|
chrX_+_102862833
|
1.686
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr5_+_68788387
|
1.683
|
NM_001205254
|
OCLN
|
occludin
|
|
chr15_+_52472257
|
1.682
|
|
LOC100129973
|
uncharacterized LOC100129973
|
|
chr17_+_60705051
|
1.666
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr5_+_112849352
|
1.658
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr4_-_170533762
|
1.658
|
NM_001199397
NM_001199398
NM_001199399
NM_001199400
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr6_+_13925423
|
1.656
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr8_+_82644663
|
1.656
|
NM_152284
|
CHMP4C
|
charged multivesicular body protein 4C
|
|
chr1_-_70671160
|
1.653
|
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr3_+_186739643
|
1.652
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr7_-_19157262
|
1.648
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr1_-_228645517
|
1.639
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr11_-_61584228
|
1.624
|
|
FADS1
|
fatty acid desaturase 1
|
|
chr6_-_121655606
|
1.623
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr6_-_43477980
|
1.601
|
|
LRRC73
|
leucine rich repeat containing 73
|
|
chr19_+_50979788
|
1.595
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr12_-_92539614
|
1.593
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr8_-_71581354
|
1.570
|
|
LACTB2
|
lactamase, beta 2
|
|
chr1_-_15850692
|
1.568
|
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr6_+_89791510
|
1.554
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr1_-_115124211
|
1.553
|
NM_005872
|
BCAS2
|
breast carcinoma amplified sequence 2
|
|
chr4_-_170533536
|
1.549
|
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr3_+_10068097
|
1.546
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr1_+_228645807
|
1.540
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chrX_+_84498993
|
1.537
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr2_+_24299751
|
1.536
|
|
LOC375190
|
UPF0638 protein B
|
|
chr18_+_57567191
|
1.535
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr12_-_6772252
|
1.529
|
NM_001127582
NM_001127583
NM_001127584
NM_001127585
NM_001127586
NM_016162
|
ING4
|
inhibitor of growth family, member 4
|
|
chr2_+_28615668
|
1.526
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr12_+_72233486
|
1.525
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr2_-_110962539
|
1.515
|
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr14_-_55493770
|
1.511
|
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr13_-_101327102
|
1.508
|
NM_001079669
NM_032813
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr14_-_95786095
|
1.505
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr19_+_36630999
|
1.501
|
|
CAPNS1
|
calpain, small subunit 1
|
|
chr19_+_36631001
|
1.488
|
|
CAPNS1
|
calpain, small subunit 1
|
|
chr17_-_33288475
|
1.473
|
NM_001193529
NM_001193530
NM_006584
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2)
|
|
chr2_+_96068423
|
1.461
|
NM_016044
|
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A
|
|
chr7_-_8302141
|
1.450
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr16_-_88772732
|
1.449
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr21_-_43187265
|
1.435
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr21_-_43187197
|
1.428
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr12_-_64616032
|
1.414
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr14_-_55493804
|
1.395
|
NM_001008396
NM_007086
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr7_+_20370324
|
1.384
|
|
ITGB8
|
integrin, beta 8
|
|
chr19_-_4517715
|
1.379
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr19_-_50980002
|
1.364
|
NM_138411
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr15_-_48470453
|
1.363
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr19_-_36499583
|
1.331
|
NM_001039876
|
C19orf46
|
chromosome 19 open reading frame 46
|
|
chr7_+_107204402
|
1.327
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr4_-_84255895
|
1.326
|
NM_001199830
|
HPSE
|
heparanase
|
|
chr22_+_38597938
|
1.325
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr3_+_50297644
|
1.325
|
|
|
|
|
chr20_+_44637539
|
1.319
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr8_+_94929082
|
1.315
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr17_+_33914278
|
1.311
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr1_-_202896368
|
1.309
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr1_+_119683006
|
1.305
|
|
|
|
|
chr12_+_133707212
|
1.303
|
NM_015394
|
ZNF10
|
zinc finger protein 10
|
|
chr5_-_178054072
|
1.296
|
|
CLK4
|
CDC-like kinase 4
|
|
chrX_+_102024057
|
1.296
|
|
LOC100287765
|
uncharacterized LOC100287765
|
|
chr20_+_17949529
|
1.294
|
NM_052865
|
C20orf72
|
chromosome 20 open reading frame 72
|